Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery
- PMID: 19557165
- PMCID: PMC2696038
- DOI: 10.1371/journal.ppat.1000493
Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery
Abstract
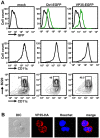
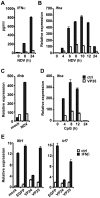
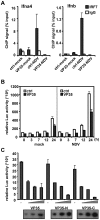
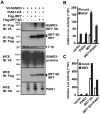
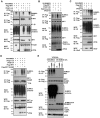
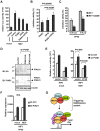
Ebola Zaire virus is highly pathogenic for humans, with case fatality rates approaching 90% in large outbreaks in Africa. The virus replicates in macrophages and dendritic cells (DCs), suppressing production of type I interferons (IFNs) while inducing the release of large quantities of proinflammatory cytokines. Although the viral VP35 protein has been shown to inhibit IFN responses, the mechanism by which it blocks IFN production has not been fully elucidated. We expressed VP35 from a mouse-adapted variant of Ebola Zaire virus in murine DCs by retroviral gene transfer, and tested for IFN transcription upon Newcastle Disease virus (NDV) infection and toll-like receptor signaling. We found that VP35 inhibited IFN transcription in DCs following these stimuli by disabling the activity of IRF7, a transcription factor required for IFN transcription. By yeast two-hybrid screens and coimmunoprecipitation assays, we found that VP35 interacted with IRF7, Ubc9 and PIAS1. The latter two are the host SUMO E2 enzyme and E3 ligase, respectively. VP35, while not itself a SUMO ligase, increased PIAS1-mediated SUMOylation of IRF7, and repressed Ifn transcription. In contrast, VP35 did not interfere with the activation of NF-kappaB, which is required for induction of many proinflammatory cytokines. Our findings indicate that Ebola Zaire virus exploits the cellular SUMOylation machinery for its advantage and help to explain how the virus overcomes host innate defenses, causing rapidly overwhelming infection to produce a syndrome resembling fulminant septic shock.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Feldmann H, Jones S, Klenk HD, Schnittler HJ. Ebola virus: from discovery to vaccine. Nat Rev Immunol. 2003;3:677–685. - PubMed
-
- Bray M, Geisbert TW. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol. 2005;37:1560–1566. - PubMed
-
- Bosio CM, Aman MJ, Grogan C, Hogan R, Ruthel G, et al. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188:1630–1638. - PubMed
-
- Mahanty S, Hutchinson K, Agarwal S, McRae M, Rollin PE, et al. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J Immunol. 2003;170:2797–2801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

