Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes
- PMID: 19557195
- PMCID: PMC2695001
- DOI: 10.1371/journal.pgen.1000536
Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes
Abstract
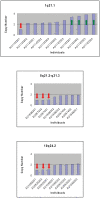
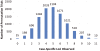
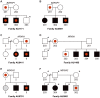
The genetics underlying the autism spectrum disorders (ASDs) is complex and remains poorly understood. Previous work has demonstrated an important role for structural variation in a subset of cases, but has lacked the resolution necessary to move beyond detection of large regions of potential interest to identification of individual genes. To pinpoint genes likely to contribute to ASD etiology, we performed high density genotyping in 912 multiplex families from the Autism Genetics Resource Exchange (AGRE) collection and contrasted results to those obtained for 1,488 healthy controls. Through prioritization of exonic deletions (eDels), exonic duplications (eDups), and whole gene duplication events (gDups), we identified more than 150 loci harboring rare variants in multiple unrelated probands, but no controls. Importantly, 27 of these were confirmed on examination of an independent replication cohort comprised of 859 cases and an additional 1,051 controls. Rare variants at known loci, including exonic deletions at NRXN1 and whole gene duplications encompassing UBE3A and several other genes in the 15q11-q13 region, were observed in the course of these analyses. Strong support was likewise observed for previously unreported genes such as BZRAP1, an adaptor molecule known to regulate synaptic transmission, with eDels or eDups observed in twelve unrelated cases but no controls (p = 2.3x10(-5)). Less is known about MDGA2, likewise observed to be case-specific (p = 1.3x10(-4)). But, it is notable that the encoded protein shows an unexpectedly high similarity to Contactin 4 (BLAST E-value = 3x10(-39)), which has also been linked to disease. That hundreds of distinct rare variants were each seen only once further highlights complexity in the ASDs and points to the continued need for larger cohorts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. - PubMed
-
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30:405–416. - PubMed
-
- Vorstman JA, Staal WG, van Daalen E, van Engeland H, Hochstenbach PF, et al. Identification of novel autism candidate regions through analysis of reported cytogenetic abnormalities associated with autism. Mol Psychiatry. 2006;11:18–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24 MH081810/MH/NIMH NIH HHS/United States
- Z01 AG000949/ImNIH/Intramural NIH HHS/United States
- 1 Z01 AG000949-02/AG/NIA NIH HHS/United States
- 1U24MH081810/MH/NIMH NIH HHS/United States
- P50 HD055784/HD/NICHD NIH HHS/United States
- R01 MH081754/MH/NIMH NIH HHS/United States
- P20-GM69012/GM/NIGMS NIH HHS/United States
- UL1 RR024134/RR/NCRR NIH HHS/United States
- MH081754/MH/NIMH NIH HHS/United States
- K24 AG000949/AG/NIA NIH HHS/United States
- P20 GM069012/GM/NIGMS NIH HHS/United States
- R01MH604687/MH/NIMH NIH HHS/United States
- UL1-RR024134-03/RR/NCRR NIH HHS/United States
- P50HD055784-01/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

