Uptake of foreign nucleic acids in kidney tubular epithelial cells deficient in proapoptotic endonucleases
- PMID: 19558214
- PMCID: PMC2903456
- DOI: 10.1089/dna.2008.0850
Uptake of foreign nucleic acids in kidney tubular epithelial cells deficient in proapoptotic endonucleases
Abstract
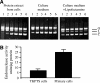
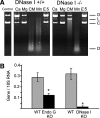
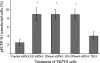
Degradation of DNA during gene delivery is an obstacle for gene transfer and for gene therapy. DNases play a major role in degrading foreign DNA. However, which of the DNases are involved and whether their inactivation can improve gene delivery have not been studied. We have recently identified deoxyribonuclease I (DNase I) and endonuclease G (EndoG) as the major degradative enzymes in the mouse kidney proximal tubule epithelial (TKPTS) cells. In this study, we used immortalized mouse TKPTS cells and primary tubular epithelial cells isolated from DNase I or EndoG knockout (KO) mice and examined the degradation of plasmid DNA during its uptake. DNase I and EndoG KO cells showed a higher rate of transfection by pECFP-N1 plasmid than wild-type cells. In addition, EndoG KO cells prevented the uptake of fluorescent-labeled RNA. Complete inhibition of secreted DNase I by G-actin did not improve plasmid transfection, indicating that only intracellular DNase I affects DNA stability. Data demonstrate the importance of DNase I and EndoG in host cell defense against gene and RNA delivery to renal tubular epithelial cells in vitro.
Figures
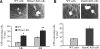


References
-
- Basnakian A.G. Apostolov E.O. Yin X. Napirei M. Mannherz H.G. Shah S.V. Cisplatin nephrotoxicity is mediated by deoxyribonuclease I. J Am Soc Nephrol. 2005;16:697–702. - PubMed
-
- Basnakian A.G. Ueda N. Kaushal G.P. Mikhailova M.V. Shah S.V. DNase I-like endonuclease in rat kidney cortex that is activated during ischemia/reperfusion injury. J Am Soc Nephrol. 2002;13:1000–1007. - PubMed
-
- Bergsmedh A. Ehnfors J. Kawane K. Motoyama N. Nagata S. Holmgren L. DNase II and the Chk2 DNA damage pathway form a genetic barrier blocking replication of horizontally transferred DNA. Mol Cancer Res. 2006;4:187–195. - PubMed
-
- Chu D. Rowe J. Lee H.C. Evaluation of the current models for the evolution of bacterial DNA uptake signal sequences. J Theor Biol. 2006;238:157–166. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

