Expressed sequence tags from larval gut of the European corn borer (Ostrinia nubilalis): exploring candidate genes potentially involved in Bacillus thuringiensis toxicity and resistance
- PMID: 19558725
- PMCID: PMC2717985
- DOI: 10.1186/1471-2164-10-286
Expressed sequence tags from larval gut of the European corn borer (Ostrinia nubilalis): exploring candidate genes potentially involved in Bacillus thuringiensis toxicity and resistance
Abstract
Background: Lepidoptera represents more than 160,000 insect species which include some of the most devastating pests of crops, forests, and stored products. However, the genomic information on lepidopteran insects is very limited. Only a few studies have focused on developing expressed sequence tag (EST) libraries from the guts of lepidopteran larvae. Knowledge of the genes that are expressed in the insect gut are crucial for understanding basic physiology of food digestion, their interactions with Bacillus thuringiensis (Bt) toxins, and for discovering new targets for novel toxins for use in pest management. This study analyzed the ESTs generated from the larval gut of the European corn borer (ECB, Ostrinia nubilalis), one of the most destructive pests of corn in North America and the western world. Our goals were to establish an ECB larval gut-specific EST database as a genomic resource for future research and to explore candidate genes potentially involved in insect-Bt interactions and Bt resistance in ECB.
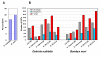
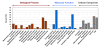
Results: We constructed two cDNA libraries from the guts of the fifth-instar larvae of ECB and sequenced a total of 15,000 ESTs from these libraries. A total of 12,519 ESTs (83.4%) appeared to be high quality with an average length of 656 bp. These ESTs represented 2,895 unique sequences, including 1,738 singletons and 1,157 contigs. Among the unique sequences, 62.7% encoded putative proteins that shared significant sequence similarities (E-value <or= 10-3)with the sequences available in GenBank. Our EST analysis revealed 52 candidate genes that potentially have roles in Bt toxicity and resistance. These genes encode 18 trypsin-like proteases, 18 chymotrypsin-like proteases, 13 aminopeptidases, 2 alkaline phosphatases and 1 cadherin-like protein. Comparisons of expression profiles of 41 selected candidate genes between Cry1Ab-susceptible and resistant strains of ECB by RT-PCR showed apparently decreased expressions in 2 trypsin-like and 2 chymotrypsin-like protease genes, and 1 aminopeptidase genes in the resistant strain as compared with the susceptible strain. In contrast, the expression of 3 trypsin- like and 3 chymotrypsin-like protease genes, 2 aminopeptidase genes, and 2 alkaline phosphatase genes were increased in the resistant strain. Such differential expressions of the candidate genes may suggest their involvement in Cry1Ab resistance. Indeed, certain trypsin-like and chymotrypsin-like proteases have previously been found to activate or degrade Bt protoxins and toxins, whereas several aminopeptidases, cadherin-like proteins and alkaline phosphatases have been demonstrated to serve as Bt receptor proteins in other insect species.
Conclusion: We developed a relatively large EST database consisting of 12,519 high-quality sequences from a total of 15,000 cDNAs from the larval gut of ECB. To our knowledge, this database represents the largest gut-specific EST database from a lepidopteran pest. Our work provides a foundation for future research to develop an ECB gut-specific DNA microarray which can be used to analyze the global changes of gene expression in response to Bt protoxins/toxins and the genetic difference(s) between Bt- resistant and susceptible strains. Furthermore, we identified 52 candidate genes that may potentially be involved in Bt toxicity and resistance. Differential expressions of 15 out of the 41 selected candidate genes examined by RT-PCR, including 5 genes with apparently decreased expression and 10 with increased expression in Cry1Ab-resistant strain, may help us conclusively identify the candidate genes involved in Bt resistance and provide us with new insights into the mechanism of Cry1Ab resistance in ECB.
Figures




Similar articles
-
Identification of a novel aminopeptidase P-like gene (OnAPP) possibly involved in Bt toxicity and resistance in a major corn pest (Ostrinia nubilalis).PLoS One. 2011;6(8):e23983. doi: 10.1371/journal.pone.0023983. Epub 2011 Aug 24. PLoS One. 2011. PMID: 21887358 Free PMC article.
-
Changes in gene expression in the larval gut of Ostrinia nubilalis in Response to Bacillus thuringiensis Cry1Ab protoxin ingestion.Toxins (Basel). 2014 Apr 3;6(4):1274-94. doi: 10.3390/toxins6041274. Toxins (Basel). 2014. PMID: 24704690 Free PMC article.
-
Characterization of cDNAs encoding serine proteases and their transcriptional responses to Cry1Ab protoxin in the gut of Ostrinia nubilalis larvae.PLoS One. 2012;7(8):e44090. doi: 10.1371/journal.pone.0044090. Epub 2012 Aug 31. PLoS One. 2012. PMID: 22952884 Free PMC article.
-
Bacillus thuringiensis toxin resistance mechanisms among Lepidoptera: progress on genomic approaches to uncover causal mutations in the European corn borer, Ostrinia nubilalis.Curr Opin Insect Sci. 2016 Jun;15:70-7. doi: 10.1016/j.cois.2016.04.003. Epub 2016 Apr 16. Curr Opin Insect Sci. 2016. PMID: 27436734 Review.
-
Insect pathogens as biological control agents: Back to the future.J Invertebr Pathol. 2015 Nov;132:1-41. doi: 10.1016/j.jip.2015.07.009. Epub 2015 Jul 27. J Invertebr Pathol. 2015. PMID: 26225455 Review.
Cited by
-
Transcriptome profiling of the intoxication response of Tenebrio molitor larvae to Bacillus thuringiensis Cry3Aa protoxin.PLoS One. 2012;7(4):e34624. doi: 10.1371/journal.pone.0034624. Epub 2012 Apr 25. PLoS One. 2012. PMID: 22558093 Free PMC article.
-
Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon.BMC Genomics. 2010 May 17;11:310. doi: 10.1186/1471-2164-11-310. BMC Genomics. 2010. PMID: 20478048 Free PMC article.
-
Newly identified APN splice isoforms suggest novel splicing mechanisms may underlie circRNA circularization in moth.FEBS Open Bio. 2019 Sep;9(9):1521-1535. doi: 10.1002/2211-5463.12689. Epub 2019 Jul 26. FEBS Open Bio. 2019. PMID: 31237102 Free PMC article.
-
Identification of a novel aminopeptidase P-like gene (OnAPP) possibly involved in Bt toxicity and resistance in a major corn pest (Ostrinia nubilalis).PLoS One. 2011;6(8):e23983. doi: 10.1371/journal.pone.0023983. Epub 2011 Aug 24. PLoS One. 2011. PMID: 21887358 Free PMC article.
-
Isolation of BAC clones containing conserved genes from libraries of three distantly related moths: a useful resource for comparative genomics of Lepidoptera.J Biomed Biotechnol. 2011;2011:165894. doi: 10.1155/2011/165894. Epub 2010 Nov 25. J Biomed Biotechnol. 2011. PMID: 21127704 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

