Gene copy number and malaria biology
- PMID: 19559648
- PMCID: PMC2839409
- DOI: 10.1016/j.pt.2009.04.005
Gene copy number and malaria biology
Abstract
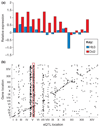
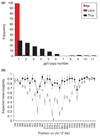
Alteration in gene copy number provides a simple way to change expression levels and alter phenotype. This was fully appreciated by bacteriologists more than 25 years ago, but the extent and implications of copy number polymorphism (CNP) have only recently become apparent in other organisms. New methods demonstrate the ubiquity of CNPs in eukaryotes and their medical importance in humans. CNP is also widespread in the Plasmodium falciparum genome and has an important and underappreciated role in determining phenotype. In this review, we summarize the distribution of CNP, its evolutionary dynamics within populations, its functional importance and its mode of evolution.
Figures



References
-
- Cooper GM, et al. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 2007;39:S22–S29. - PubMed
-
- Freeman JL, et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. - PubMed
-
- McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. - PubMed
-
- Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

