Mechanical constraints on Hin subunit rotation imposed by the Fis/enhancer system and DNA supercoiling during site-specific recombination
- PMID: 19560425
- PMCID: PMC2752211
- DOI: 10.1016/j.molcel.2009.05.020
Mechanical constraints on Hin subunit rotation imposed by the Fis/enhancer system and DNA supercoiling during site-specific recombination
Abstract
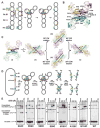
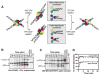
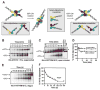
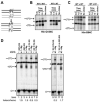
Hin, a member of the serine family of site-specific recombinases, regulates gene expression by inverting a DNA segment. DNA inversion requires assembly of an invertasome complex in which a recombinational enhancer DNA segment bound by the Fis protein associates with the Hin synaptic complex at the base of a supercoiled DNA branch. Each of the four Hin subunits becomes covalently joined to the cleaved DNA ends, and DNA exchange occurs by translocation of a Hin subunit pair within the tetramer. We show here that, although the Hin tetramer forms a bidirectional molecular swivel, the Fis/enhancer system determines both the direction and number of subunit rotations. The chirality of supercoiling directs rotational direction, and the short DNA loop stabilized by Fis-Hin contacts limit rotational processivity, thereby ensuring that the DNA strands religate in the recombinant configuration. We identify multiple rotational conformers that are formed under different supercoiling and solution conditions.
Figures



Similar articles
-
Multiple interfaces between a serine recombinase and an enhancer control site-specific DNA inversion.Elife. 2013 Oct 22;2:e01211. doi: 10.7554/eLife.01211. Elife. 2013. PMID: 24151546 Free PMC article.
-
Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction.EMBO J. 1989 May;8(5):1581-90. doi: 10.1002/j.1460-2075.1989.tb03542.x. EMBO J. 1989. PMID: 2548848 Free PMC article.
-
The Hin dimer interface is critical for Fis-mediated activation of the catalytic steps of site-specific DNA inversion.Curr Biol. 1996 Feb 1;6(2):163-77. doi: 10.1016/s0960-9822(02)00449-9. Curr Biol. 1996. PMID: 8673463
-
Site-specific DNA Inversion by Serine Recombinases.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0047-2014. doi: 10.1128/microbiolspec.MDNA3-0047-2014. Microbiol Spectr. 2015. PMID: 26104558 Review.
-
DNA inversions in phages and bacteria.Trends Genet. 1992 Dec;8(12):457-62. doi: 10.1016/0168-9525(92)90331-w. Trends Genet. 1992. PMID: 1337227 Review.
Cited by
-
Single-Molecule Tethered Particle Motion: Stepwise Analyses of Site-Specific DNA Recombination.Micromachines (Basel). 2018 May 3;9(5):216. doi: 10.3390/mi9050216. Micromachines (Basel). 2018. PMID: 30424148 Free PMC article. Review.
-
Sin resolvase catalytic activity and oligomerization state are tightly coupled.J Mol Biol. 2010 Nov 19;404(1):16-33. doi: 10.1016/j.jmb.2010.08.057. Epub 2010 Sep 22. J Mol Biol. 2010. PMID: 20868695 Free PMC article.
-
Direct observation of subunit rotation during DNA strand exchange by serine recombinases.Nat Commun. 2024 Nov 29;15(1):10407. doi: 10.1038/s41467-024-54531-4. Nat Commun. 2024. PMID: 39613732 Free PMC article.
-
Effect of iacP mutation on flagellar phase variation in Salmonella enterica serovar typhimurium strain UK-1.J Bacteriol. 2012 Aug;194(16):4332-41. doi: 10.1128/JB.00076-12. Epub 2012 Jun 8. J Bacteriol. 2012. PMID: 22685287 Free PMC article.
-
Multiple interfaces between a serine recombinase and an enhancer control site-specific DNA inversion.Elife. 2013 Oct 22;2:e01211. doi: 10.7554/eLife.01211. Elife. 2013. PMID: 24151546 Free PMC article.
References
-
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. - PubMed
-
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, D.C.: ASM Press; 2002.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

