Shifts in replication timing actively affect histone acetylation during nucleosome reassembly
- PMID: 19560427
- PMCID: PMC2717743
- DOI: 10.1016/j.molcel.2009.05.027
Shifts in replication timing actively affect histone acetylation during nucleosome reassembly
Abstract
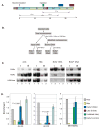
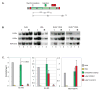
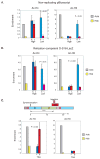
The entire genome is replicated in a programmed manner, with specific regions undergoing DNA synthesis at different times in S phase. Active genes generally replicate in early S phase, while repressed genes replicate late, and for some loci this process is developmentally regulated. Using a nuclear microinjection system, we demonstrate that DNA sequences originally packaged into nucleosomes containing deacetylated histones during late S become reassembled with acetylated histones after undergoing replication in early S. Conversely, a change from early to late replication timing is accompanied by repackaging into nucleosomes containing deacetylated histones. This is carried out by differential cell-cycle-controlled acetylation and deacetylation of histones H3 and H4. These studies provide strong evidence that switches in replication timing may play a role in the regulation of nucleosome structure during development.
Figures

References
-
- Allshire R, Bickmore W. Pausing for thought on the boundaries of imprinting. Cell. 2000;102:705–708. - PubMed
-
- Azuara V, Brown KE, Williams RR, Webb N, Dillon N, Festenstein R, Buckle V, Merkenschlager M, Fisher AG. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nature Cell Biol. 2003;5:668–674. - PubMed
-
- Brown DD. The role of stable complexes that repress and activate eucaryotic genes. Cell. 1984;37:359–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

