The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor
- PMID: 19561086
- PMCID: PMC2782041
- DOI: 10.1074/jbc.M109.025007
The small terminase, gp16, of bacteriophage T4 is a regulator of the DNA packaging motor
Abstract
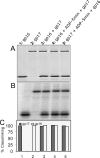
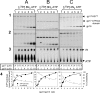
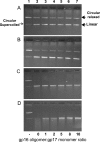
Tailed bacteriophages and herpes viruses use powerful molecular motors to translocate DNA into a preassembled prohead and compact the DNA to near crystalline density. The phage T4 motor, a pentamer of 70-kDa large terminase, gp17, is the fastest and most powerful motor reported to date. gp17 has an ATPase activity that powers DNA translocation and a nuclease activity that cuts concatemeric DNA and generates the termini of viral genome. An 18-kDa small terminase, gp16, is also essential, but its role in DNA packaging is poorly understood. gp16 forms oligomers, most likely octamers, exhibits no enzymatic activities, but stimulates the gp17-ATPase activity, and inhibits the nuclease activity. Extensive mutational and biochemical analyses show that gp16 contains three domains, a central oligomerization domain, and N- and C-terminal domains that are essential for ATPase stimulation. Stimulation occurs not by nucleotide exchange or enhanced ATP binding but by triggering hydrolysis of gp17-bound ATP, a mechanism reminiscent of GTPase-activating proteins. gp16 does not have an arginine finger but its interaction with gp17 seems to position a gp17 arginine finger into the catalytic pocket. gp16 inhibits DNA translocation when gp17 is associated with the prohead. gp16 restricts gp17-nuclease such that the putative packaging initiation cut is made but random cutting is inhibited. These results suggest that the phage T4 packaging machine consists of a motor (gp17) and a regulator (gp16). The gp16 regulator is essential to coordinate the gp17 motor ATPase, translocase, and nuclease activities, otherwise it could be suicidal to the virus.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

