FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures
- PMID: 19561358
- PMCID: PMC2782037
- DOI: 10.1074/jbc.M109.016006
FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures
Abstract
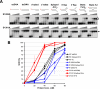
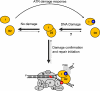
In this study, we report that the purified wild-type FANCI (Fanconi anemia complementation group I) protein directly binds to a variety of DNA substrates. The DNA binding domain roughly encompasses residues 200-1000, as suggested by the truncation study. When co-expressed in insect cells, a small fraction of FANCI forms a stable complex with FANCD2 (Fanconi anemia complementation group D2). Intriguingly, the purified FANCI-FANCD2 complex preferentially binds to the branched DNA structures when compared with either FANCI or FANCD2 alone. Co-immunoprecipitation with purified proteins indicates that FANCI interacts with FANCD2 through its C-terminal amino acid 1001-1328 fragment. Although the C terminus of FANCI is dispensable for direct DNA binding, it seems to be involved in the regulation of DNA binding activity. This notion is further enhanced by two C-terminal point mutations, R1285Q and D1301A, which showed differentiated DNA binding activity. We also demonstrate that FANCI forms discrete nuclear foci in HeLa cells in the absence or presence of exogenous DNA damage. The FANCI foci are colocalized perfectly with FANCD2 and partially with proliferating cell nuclear antigen irrespective of mitomycin C treatment. An increased number of FANCI foci form and become resistant to Triton X extraction in response to mitomycin C treatment. Our data suggest that the FANCI-FANCD2 complex may participate in repair of damaged replication forks through its preferential recognition of branched structures.
Figures




References
-
- Alter B. P., Greene M. H., Velazquez I., Rosenberg P. S. (2003) Blood 101, 2072. - PubMed
-
- de Winter J. P., Joenje H. (2009) Mutat. Res. 668, 11–19 - PubMed
-
- Joenje H., Patel K. J. (2001) Nat. Rev. Genet. 2, 446–457 - PubMed
-
- Kennedy R. D., D'Andrea A. D. (2005) Genes Dev. 19, 2925–2940 - PubMed
-
- Kutler D. I., Singh B., Satagopan J., Batish S. D., Berwick M., Giampietro P. F., Hanenberg H., Auerbach A. D. (2003) Blood 101, 1249–1256 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

