The interaction of mitochondrial iron with manganese superoxide dismutase
- PMID: 19561359
- PMCID: PMC2755670
- DOI: 10.1074/jbc.M109.026773
The interaction of mitochondrial iron with manganese superoxide dismutase
Abstract
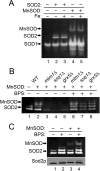
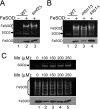
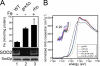
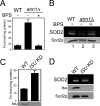
Superoxide dismutase 2 (SOD2) is one of the rare mitochondrial enzymes evolved to use manganese as a cofactor over the more abundant element iron. Although mitochondrial iron does not normally bind SOD2, iron will misincorporate into Saccharomyces cerevisiae Sod2p when cells are starved for manganese or when mitochondrial iron homeostasis is disrupted by mutations in yeast grx5, ssq1, and mtm1. We report here that such changes in mitochondrial manganese and iron similarly affect cofactor selection in a heterologously expressed Escherichia coli Mn-SOD, but not a highly homologous Fe-SOD. By x-ray absorption near edge structure and extended x-ray absorption fine structure analyses of isolated mitochondria, we find that misincorporation of iron into yeast Sod2p does not correlate with significant changes in the average oxidation state or coordination chemistry of bulk mitochondrial iron. Instead, small changes in mitochondrial iron are likely to promote iron-SOD2 interactions. Iron binds Sod2p in yeast mutants blocking late stages of iron-sulfur cluster biogenesis (grx5, ssq1, and atm1), but not in mutants defective in the upstream Isu proteins that serve as scaffolds for iron-sulfur biosynthesis. In fact, we observed a requirement for the Isu proteins in iron inactivation of yeast Sod2p. Sod2p activity was restored in mtm1 and grx5 mutants by depleting cells of Isu proteins or using a dominant negative Isu1p predicted to stabilize iron binding to Isu1p. In all cases where disruptions in iron homeostasis inactivated Sod2p, we observed an increase in mitochondrial Isu proteins. These studies indicate that the Isu proteins and the iron-sulfur pathway can donate iron to Sod2p.
Figures
References
-
- Wintjens R., Noël C., May A. C., Gerbod D., Dufernez F., Capron M., Viscogliosi E., Rooman M. (2004) J. Biol. Chem. 279,9248–9254 - PubMed
-
- Carlioz A., Ludwig M. L., Stallings W. C., Fee J. A., Steinman H. M., Touati D. (1988) J. Biol. Chem. 263,1555–1562 - PubMed
-
- Whittaker J. W. (2003) Biochem. Soc. Trans. 31,1318–1321 - PubMed
-
- Vance C. K., Miller A. F. (1998) J. Am. Chem. Soc. 120,461–467
-
- Vance C. K., Miller A. F. (2001) Biochemistry 40,13079–13087 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

