Use with caution: developmental systems divergence and potential pitfalls of animal models
- PMID: 19562005
- PMCID: PMC2701150
Use with caution: developmental systems divergence and potential pitfalls of animal models
Abstract
Transgenic animal models have played an important role in elucidating gene functions and the molecular basis development, physiology, behavior, and pathogenesis. Transgenic models have been so successful that they have become a standard tool in molecular genetics and biomedical studies and are being used to fulfill one of the main goals of the post-genomic era: to assign functions to each gene in the genome. However, the assumption that gene functions and genetic systems are conserved between models and humans is taken for granted, often in spite of evidence that gene functions and networks diverge during evolution. In this review, I discuss some mechanisms that generate functional divergence and highlight recent examples demonstrating that gene functions and regulatory networks diverge through time. These examples suggest that annotation of gene functions based solely on mutant phenotypes in animal models, as well as assumptions of conserved functions between species, can be wrong. Therefore, animal models of gene function and human disease may not provide appropriate information, particularly for rapidly evolving genes and systems.
Figures
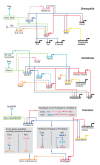

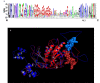
Similar articles
-
Inference of Developmental Gene Regulatory Networks Beyond Classical Model Systems: New Approaches in the Post-genomic Era.Integr Comp Biol. 2018 Oct 1;58(4):640-653. doi: 10.1093/icb/icy061. Integr Comp Biol. 2018. PMID: 29917089 Review.
-
Genome-wide pathogenesis interpretation using a heat diffusion-based systems genetics method and implications for gene function annotation.Mol Genet Genomic Med. 2020 Oct;8(10):e1456. doi: 10.1002/mgg3.1456. Epub 2020 Sep 1. Mol Genet Genomic Med. 2020. PMID: 32869547 Free PMC article.
-
Genetic evidence for conserved non-coding element function across species-the ears have it.Front Physiol. 2014 Jan 21;5:7. doi: 10.3389/fphys.2014.00007. eCollection 2014. Front Physiol. 2014. PMID: 24478720 Free PMC article.
-
Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis.BMC Genomics. 2013 Aug 20;14:568. doi: 10.1186/1471-2164-14-568. BMC Genomics. 2013. PMID: 23961710 Free PMC article.
-
Genetically altered mouse models: the good, the bad, and the ugly.Crit Rev Oral Biol Med. 2003;14(3):154-74. doi: 10.1177/154411130301400302. Crit Rev Oral Biol Med. 2003. PMID: 12799320 Review.
Cited by
-
Tunicates: exploring the sea shores and roaming the open ocean. A tribute to Thomas Huxley.Open Biol. 2015 Jun;5(6):150053. doi: 10.1098/rsob.150053. Open Biol. 2015. PMID: 26085517 Free PMC article. Review.
-
Emerging from the bottleneck: benefits of the comparative approach to modern neuroscience.Trends Neurosci. 2015 May;38(5):273-8. doi: 10.1016/j.tins.2015.02.008. Epub 2015 Mar 21. Trends Neurosci. 2015. PMID: 25800324 Free PMC article. Review.
-
Animal models in epilepsy research: legacies and new directions.Nat Neurosci. 2015 Mar;18(3):339-43. doi: 10.1038/nn.3934. Epub 2015 Feb 24. Nat Neurosci. 2015. PMID: 25710835 Review.
-
Can prospective systematic reviews of animal studies improve clinical translation?J Transl Med. 2020 Jan 9;18(1):15. doi: 10.1186/s12967-019-02205-x. J Transl Med. 2020. PMID: 31918734 Free PMC article.
-
Pitfalls of exome sequencing: a case study of the attribution of HABP2 rs7080536 in familial non-medullary thyroid cancer.NPJ Genom Med. 2017;2:8. doi: 10.1038/s41525-017-0011-x. Epub 2017 Mar 28. NPJ Genom Med. 2017. PMID: 28884020 Free PMC article.
References
-
- Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swims into view. Nature Reviews Genetics. 2007;8:250–267. - PubMed
-
- Rosenthal N, Brown S. The mouse ascending: perspectives for human-disease models. Nat Cell Biol. 2007;9(9):993–999. - PubMed
-
- Hau J. Animal models for human disease. Berlin, Germany: Humana Press Inc.; 2008.
-
- Koonin EV. Orthologs, paralogs and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. - PubMed
-
- Amundson R. The changing role of the embryo in evolutionary thought: roots of Evo-Devo. Cambridge: Cambridge University Press; 2005.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
