A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway
- PMID: 19562034
- PMCID: PMC2698153
- DOI: 10.1371/journal.pone.0006068
A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway
Abstract
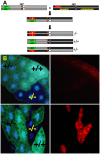
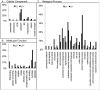
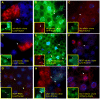
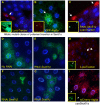
The highly conserved autophagy-lysosome pathway is the primary mechanism for breakdown and recycling of macromolecular and organellar cargo in the eukaryotic cell. Autophagy has recently been implicated in protection against cancer, neurodegeneration, and infection, and interest is increasing in additional roles of autophagy in human health, disease, and aging. To search for novel cytoprotective features of this pathway, we carried out a genetic mosaic screen for mutations causing increased lysosomal and/or autophagic activity in the Drosophila melanogaster larval fat body. By combining Drosophila genetics with live-cell imaging of the fluorescent dye LysoTracker Red and fixed-cell imaging of autophagy-specific fluorescent protein markers, the screen was designed to identify essential metazoan genes whose disruption causes increased flux through the autophagy-lysosome pathway. The screen identified a large number of genes associated with the protein synthesis and ER-secretory pathways (e.g. aminoacyl tRNA synthetases, Oligosaccharyl transferase, Sec61alpha), and with mitochondrial function and dynamics (e.g. Rieske iron-sulfur protein, Dynamin-related protein 1). We also observed that increased lysosomal and autophagic activity were consistently associated with decreased cell size. Our work demonstrates that disruption of the synthesis, transport, folding, or glycosylation of ER-targeted proteins at any of multiple steps leads to autophagy induction. In addition to illuminating cytoprotective features of autophagy in response to cellular damage, this screen establishes a genetic methodology for investigating cell biological phenotypes in live cells, in the context of viable wild type organisms.
Conflict of interest statement
Figures



References
-
- Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
-
- Vellodi A. Lysosomal storage disorders. Br J Haematol. 2005;128:413–431. - PubMed
-
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

