Pharmacogenetics and population pharmacokinetics: impact of the design on three tests using the SAEM algorithm
- PMID: 19562469
- PMCID: PMC3192406
- DOI: 10.1007/s10928-009-9124-x
Pharmacogenetics and population pharmacokinetics: impact of the design on three tests using the SAEM algorithm
Abstract
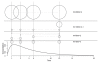
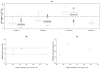
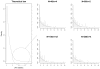
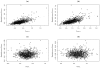
Pharmacogenetics is now widely investigated and health institutions acknowledge its place in clinical pharmacokinetics. Our objective is to assess through a simulation study, the impact of design on the statistical performances of three different tests used for analysis of pharmacogenetic information with nonlinear mixed effects models: (i) an ANOVA to test the relationship between the empirical Bayes estimates of the model parameter of interest and the genetic covariate, (ii) a global Wald test to assess whether estimates for the gene effect are significant, and (iii) a likelihood ratio test (LRT) between the model with and without the genetic covariate. We use the stochastic EM algorithm (SAEM) implemented in MONOLIX 2.1 software. The simulation setting is inspired from a real pharmacokinetic study. We investigate four designs with N the number of subjects and n the number of samples per subject: (i) N = 40/n = 4, similar to the original study, (ii) N = 80/n = 2 sorted in 4 groups, a design optimized using the PFIM software, (iii) a combined design, N = 20/n = 4 plus N = 80 with only a trough concentration and (iv) N = 200/n = 4, to approach asymptotic conditions. We find that the ANOVA has a correct type I error estimate regardless of design, however the sparser design was optimized. The type I error of the Wald test and LRT are moderatly inflated in the designs far from the asymptotic (<10%). For each design, the corrected power is analogous for the three tests. Among the three designs with a total of 160 observations, the design N = 80/n = 2 optimized with PFIM provides both the lowest standard error on the effect coefficients and the best power for the Wald test and the LRT while a high shrinkage decreases the power of the ANOVA. In conclusion, a correction method should be used for model-based tests in pharmacogenetic studies with reduced sample size and/or sparse sampling and, for the same amount of samples, some designs have better power than others.
Figures


References
-
- EMEA. Technical report. EMEA; 2008. ICH topic E15 definitions for genomic biomarkers, pharmacogenomics, pharmacogenetics, genomic data and sample coding categories.
-
- FDA. Technical report. FDA; 2008. E15 definitions for genomic biomarkers, pharmacogenomics, pharmacogenetics, genomic data and sample coding categories.
-
- EMEA. Technical report. EMEA; 2007. Reflection paper on the use of pharmacogenetics in the pharmacokinetic evaluation of medicinal products.
-
- Hu XP, Xu JM, Hu YM, Mei Q, Xu XH. Effects of CYP2C19 genetic polymorphism on the pharmacokinetics and pharmacodynamics of omeprazole in chinese people. J Clin Pharmacol Ther. 2007;32:517–524. - PubMed

