Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism
- PMID: 19563756
- PMCID: PMC3710970
- DOI: 10.1016/j.cell.2009.04.024
Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism
Abstract
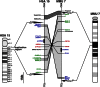
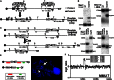
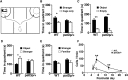
Substantial evidence suggests that chromosomal abnormalities contribute to the risk of autism. The duplication of human chromosome 15q11-13 is known to be the most frequent cytogenetic abnormality in autism. We have modeled this genetic change in mice by using chromosome engineering to generate a 6.3 Mb duplication of the conserved linkage group on mouse chromosome 7. Mice with a paternal duplication display poor social interaction, behavioral inflexibility, abnormal ultrasonic vocalizations, and correlates of anxiety. An increased MBII52 snoRNA within the duplicated region, affecting the serotonin 2c receptor (5-HT2cR), correlates with altered intracellular Ca(2+) responses elicited by a 5-HT2cR agonist in neurons of mice with a paternal duplication. This chromosome-engineered mouse model for autism seems to replicate various aspects of human autistic phenotypes and validates the relevance of the human chromosome abnormality. This model will facilitate forward genetics of developmental brain disorders and serve as an invaluable tool for therapeutic development.
Figures



References
-
- Akashi M., Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005;12:441–448. - PubMed
-
- Beaudet A.L. Autism: highly heritable but not inherited. Nat. Med. 2007;13:534–536. - PubMed
-
- Belmonte M.K., Cook E.H., Jr., Anderson G.M., Rubenstein J.L., Greenough W.T., Beckel-Mitchener A., Courchesne E., Boulanger L.M., Powell S.B., Levitt P.R. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol. Psychiatry. 2004;9:646–663. - PubMed
-
- Bolton P.F., Veltman M.W., Weisblatt E., Holmes J.R., Thomas N.S., Youings S.A., Thompson R.J., Roberts S.E., Dennis N.R., Browne C.E. Chromosome 15q11-13 abnormalities and other medical conditions in individuals with autism spectrum disorders. Psychiatr. Genet. 2004;14:131–137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

