Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment
- PMID: 19563760
- PMCID: PMC2814540
- DOI: 10.1016/j.cell.2009.04.025
Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment
Erratum in
- Cell. 2009 Aug 7;138(3):604
Abstract
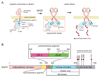
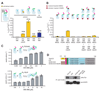
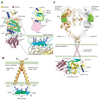
Signaling by the epidermal growth factor receptor requires an allosteric interaction between the kinase domains of two receptors, whereby one activates the other. We show that the intracellular juxtamembrane segment of the receptor, known to potentiate kinase activity, is able to dimerize the kinase domains. The C-terminal half of the juxtamembrane segment latches the activated kinase domain to the activator, and the N-terminal half of this segment further potentiates dimerization, most likely by forming an antiparallel helical dimer that engages the transmembrane helices of the activated receptor. Our data are consistent with a mechanism in which the extracellular domains block the intrinsic ability of the transmembrane and cytoplasmic domains to dimerize and activate, with ligand binding releasing this block. The formation of the activating juxtamembrane latch is prevented by the C-terminal tails in a structure of an inactive kinase domain dimer, suggesting how alternative dimers can prevent ligand-independent activation.
Figures




Comment in
-
The juxtamembrane region of EGFR takes center stage.Cell. 2009 Jun 26;137(7):1181-3. doi: 10.1016/j.cell.2009.06.008. Cell. 2009. PMID: 19563749
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. - PubMed
-
- Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J Biol Chem. 2008;283:6950–6956. - PubMed
-
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. - PubMed
-
- Burke CL, Lemmon MA, Coren BA, Engelman DM, Stern DF. Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene. 1997;14:687–696. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

