Peripheral deletion of self-reactive B cells
- PMID: 1956380
- PMCID: PMC3787863
- DOI: 10.1038/354308a0
Peripheral deletion of self-reactive B cells
Abstract
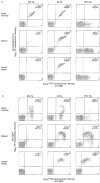
B LYMPHOCYTES are key participants in the immune response because of their specificity, their ability to take up and present antigens to T cells, and their capacity to differentiate into antibody-secreting cells. To limit reactivity to self antigens, autospecific B cells can be functionally inactivated or deleted. Developing B cells that react with membrane antigens expressed in the bone marrow are deleted from the peripheral lymphocyte pool. It is important to ascertain the fate of B cells that recognize membrane autoantigens expressed exclusively on peripheral tissues because B cells in the peripheral lymphoid organs are phenotypically and functionally distinct from bone-marrow B cells. Here we show that in immunoglobulin-transgenic mice, B cells specific for major histocompatibility complex class I antigen can be deleted if they encounter membrane-bound antigen at a post-bone-marrow stage of development. This deletion may be necessary to prevent organ-specific autoimmunity.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

