Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine
- PMID: 19564383
- PMCID: PMC2738016
- DOI: 10.1128/IAI.00279-09
Sialic acid catabolism confers a competitive advantage to pathogenic vibrio cholerae in the mouse intestine
Abstract
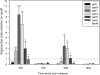
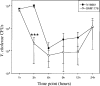
Sialic acids comprise a family of nine-carbon ketosugars that are ubiquitous on mammalian mucous membranes. However, sialic acids have a limited distribution among Bacteria and are confined mainly to pathogenic and commensal species. Vibrio pathogenicity island 2 (VPI-2), a 57-kb region found exclusively among pathogenic strains of Vibrio cholerae, contains a cluster of genes (nan-nag) putatively involved in the scavenging (nanH), transport (dctPQM), and catabolism (nanA, nanE, nanK, and nagA) of sialic acid. The capacity to utilize sialic acid as a carbon and energy source might confer an advantage to V. cholerae in the mucus-rich environment of the gut, where sialic acid availability is extensive. In this study, we show that V. cholerae can utilize sialic acid as a sole carbon source. We demonstrate that the genes involved in the utilization of sialic acid are located within the nan-nag region of VPI-2 by complementation of Escherichia coli mutants and gene knockouts in V. cholerae N16961. We show that nanH, dctP, nanA, and nanK are highly expressed in V. cholerae grown on sialic acid. By using the infant mouse model of infection, we show that V. cholerae DeltananA strain SAM1776 is defective in early intestinal colonization stages. In addition, SAM1776 shows a decrease in the competitive index in colonization-competition assays comparing the mutant strain with both O1 El Tor and classical strains. Our data indicate an important relationship between the catabolism of sialic acid and bacterial pathogenesis, stressing the relevance of the utilization of the resources found in the host's environment.
Figures
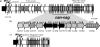


References
-
- Alam, M., N. A. Hasan, A. Sadique, N. A. Bhuiyan, K. U. Ahmed, S. Nusrin, G. B. Nair, A. K. Siddique, R. B. Sack, D. A. Sack, A. Huq, and R. R. Colwell. 2006. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl. Environ. Microbiol. 724096-4104. - PMC - PubMed
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

