A site-specific, multiplexed kinase activity assay using stable-isotope dilution and high-resolution mass spectrometry
- PMID: 19564600
- PMCID: PMC2710625
- DOI: 10.1073/pnas.0905165106
A site-specific, multiplexed kinase activity assay using stable-isotope dilution and high-resolution mass spectrometry
Abstract
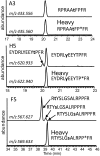
Most kinases are capable of recognizing and phosphorylating peptides containing short, linear sequence motifs. To measure the activation state of many kinases from the same cell lysate, we created a multiplexed, mass-spectrometry-based in vitro kinase assay. Ninety chemically synthesized peptides derived from well-characterized peptide substrates and in vivo phosphorylation sites with either known or previously unidentified upstream kinases were reacted individually in a plate format with crude cell lysates and ATP. Phosphorylation rates were directly measured based on the addition of 90 same-sequence, site-specific phosphopeptides enriched in stable isotopes to act as ideal quantitative internal standards for analysis by liquid chromatography coupled to tandem mass spectrometry. This approach concurrently measured up to 90 site-specific peptide phosphorylation rates, reporting a diagnostic fingerprint for activated kinase pathways. We applied this unique kinome-activity profiling strategy in a variety of cellular settings, including mitogen stimulation, cell cycle, pharmacological inhibition of pathways, and to a panel of breast cancer cell lines. Finally, we identified the source of activity for a peptide (derived from a PI3K regulatory subunit) from our library. This peptide substrate demonstrated mitotic and tyrosine-specific phosphorylation, which was confirmed to be a novel Src family kinase site in vivo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5:172–183. - PubMed
-
- Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–480. - PubMed
-
- Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

