High-resolution profiling and discovery of planarian small RNAs
- PMID: 19564616
- PMCID: PMC2703670
- DOI: 10.1073/pnas.0905222106
High-resolution profiling and discovery of planarian small RNAs
Abstract
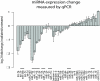
Freshwater planarian flatworms possess uncanny regenerative capacities mediated by abundant and collectively totipotent adult stem cells. Key functions of these cells during regeneration and tissue homeostasis have been shown to depend on PIWI, a molecule required for Piwi-interacting RNA (piRNA) expression in planarians. Nevertheless, the full complement of piRNAs and microRNAs (miRNAs) in this organism has yet to be defined. Here we report on the large-scale cloning and sequencing of small RNAs from the planarian Schmidtea mediterranea, yielding altogether millions of sequenced, unique small RNAs. We show that piRNAs are in part organized in genomic clusters and that they share characteristic features with mammalian and fly piRNAs. We further identify 61 novel miRNA genes and thus double the number of known planarian miRNAs. Sequencing, as well as quantitative PCR of small RNAs, uncovered 10 miRNAs enriched in planarian stem cells. These miRNAs are down-regulated in animals in which stem cells have been abrogated by irradiation, and thus constitute miRNAs likely associated with specific stem-cell functions. Altogether, we present the first comprehensive small RNA analysis in animals belonging to the third animal superphylum, the Lophotrochozoa, and single out a number of miRNAs that may function in regeneration. Several of these miRNAs are deeply conserved in animals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. - PubMed
-
- Randolph H. The regeneration of the tail in lumbriculus. J Morphol. 1892;7:317–344.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

