Evolutionary analysis of the dynamics of viral infectious disease
- PMID: 19564871
- PMCID: PMC7097015
- DOI: 10.1038/nrg2583
Evolutionary analysis of the dynamics of viral infectious disease
Abstract
Many organisms that cause infectious diseases, particularly RNA viruses, mutate so rapidly that their evolutionary and ecological behaviours are inextricably linked. Consequently, aspects of the transmission and epidemiology of these pathogens are imprinted on the genetic diversity of their genomes. Large-scale empirical analyses of the evolutionary dynamics of important pathogens are now feasible owing to the increasing availability of pathogen sequence data and the development of new computational and statistical methods of analysis. In this Review, we outline the questions that can be answered using viral evolutionary analysis across a wide range of biological scales.
Figures
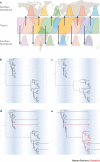
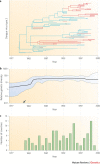
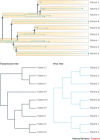
References
-
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nature Rev. Genet. 2008;9:267–276. - PubMed
-
- Grenfell BT, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. - PubMed
-
- Holmes EC, Nee S, Rambaut A, Garnett GP, Harvey PH. Revealing the history of infectious disease epidemics through phylogenetic trees. Philos. Trans. R. Soc. Lond. B. 1995;349:33–40. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

