Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection
- PMID: 19564922
- PMCID: PMC2699633
- DOI: 10.1371/journal.pone.0006093
Synergistic activation of HIV-1 expression by deacetylase inhibitors and prostratin: implications for treatment of latent infection
Abstract
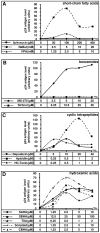
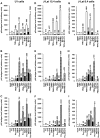
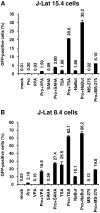
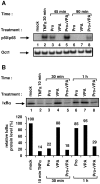
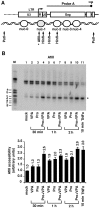
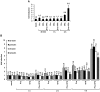
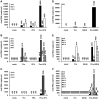
The persistence of transcriptionally silent but replication-competent HIV-1 reservoirs in Highly Active Anti-Retroviral Therapy (HAART)-treated infected individuals, represents a major hurdle to virus eradication. Activation of HIV-1 gene expression in these cells together with an efficient HAART has been proposed as an adjuvant therapy aimed at decreasing the pool of latent viral reservoirs. Using the latently-infected U1 monocytic cell line and latently-infected J-Lat T-cell clones, we here demonstrated a strong synergistic activation of HIV-1 production by clinically used histone deacetylase inhibitors (HDACIs) combined with prostratin, a non-tumor-promoting nuclear factor (NF)- kappaB inducer. In J-Lat cells, we showed that this synergism was due, at least partially, to the synergistic recruitment of unresponsive cells into the expressing cell population. A combination of prostratin+HDACI synergistically activated the 5' Long Terminal Repeat (5'LTR) from HIV-1 Major group subtypes representing the most prevalent viral genetic forms, as shown by transient transfection reporter assays. Mechanistically, HDACIs increased prostratin-induced DNA-binding activity of nuclear NF-kappaB and degradation of cytoplasmic NF-kappaB inhibitor, IkappaBalpha . Moreover, the combined treatment prostratin+HDACI caused a more pronounced nucleosomal remodeling in the U1 viral promoter region than the treatments with the compounds alone. This more pronounced remodeling correlated with a synergistic reactivation of HIV-1 transcription following the combined treatment prostratin+HDACI, as demonstrated by measuring recruitment of RNA polymerase II to the 5'LTR and both initiated and elongated transcripts. The physiological relevance of the prostratin+HDACI synergism was shown in CD8(+)-depleted peripheral blood mononuclear cells from HAART-treated patients with undetectable viral load. Moreover, this combined treatment reactivated viral replication in resting CD4(+) T cells isolated from similar patients. Our results suggest that combinations of different kinds of proviral activators may have important implications for reducing the size of latent HIV-1 reservoirs in HAART-treated patients.
Conflict of interest statement
Figures
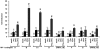
References
-
- Bisgrove D, Lewinski M, Bushman F, Verdin E. Molecular mechanisms of HIV-1 proviral latency. Expert Review of Anti-infective Therapy. 2005;3:805–814. - PubMed
-
- Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol. 2007;5:95–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

