Copy number variations and cancer
- PMID: 19566914
- PMCID: PMC2703871
- DOI: 10.1186/gm62
Copy number variations and cancer
Abstract
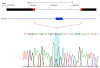
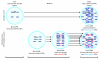
DNA copy number variations (CNVs) are an important component of genetic variation, affecting a greater fraction of the genome than single nucleotide polymorphisms (SNPs). The advent of high-resolution SNP arrays has made it possible to identify CNVs. Characterization of widespread constitutional (germline) CNVs has provided insight into their role in susceptibility to a wide spectrum of diseases, and somatic CNVs can be used to identify regions of the genome involved in disease phenotypes. The role of CNVs as risk factors for cancer is currently underappreciated. However, the genomic instability and structural dynamism that characterize cancer cells would seem to make this form of genetic variation particularly intriguing to study in cancer. Here, we provide a detailed overview of the current understanding of the CNVs that arise in the human genome and explore the emerging literature that reveals associations of both constitutional and somatic CNVs with a wide variety of human cancers.
Figures

References
-
- Gault J, Hopkins J, Berger R, Drebing C, Logel J, Walton C, Short M, Vianzon R, Olincy A, Ross RG, Adler LE, Freedman R, Leonard S. Comparison of polymorphisms in the alpha7 nicotinic receptor gene and its partial duplication in schizophrenic and control subjects. Am J Med Genet B Neuropsychiatr Genet. 2003;123B:39–49. doi: 10.1002/ajmg.b.20061. - DOI - PubMed
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

