Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease
- PMID: 19567438
- PMCID: PMC2803020
- DOI: 10.1001/jama.2009.954
Genetic Loci associated with C-reactive protein levels and risk of coronary heart disease
Abstract
Context: Plasma levels of C-reactive protein (CRP) are independently associated with risk of coronary heart disease, but whether CRP is causally associated with coronary heart disease or merely a marker of underlying atherosclerosis is uncertain.
Objective: To investigate association of genetic loci with CRP levels and risk of coronary heart disease.
Design, setting, and participants: We first carried out a genome-wide association (n = 17,967) and replication study (n = 13,615) to identify genetic loci associated with plasma CRP concentrations. Data collection took place between 1989 and 2008 and genotyping between 2003 and 2008. We carried out a mendelian randomization study of the most closely associated single-nucleotide polymorphism (SNP) in the CRP locus and published data on other CRP variants involving a total of 28,112 cases and 100,823 controls, to investigate the association of CRP variants with coronary heart disease. We compared our finding with that predicted from meta-analysis of observational studies of CRP levels and risk of coronary heart disease. For the other loci associated with CRP levels, we selected the most closely associated SNP for testing against coronary heart disease among 14,365 cases and 32,069 controls.
Main outcome measure: Risk of coronary heart disease.
Results: Polymorphisms in 5 genetic loci were strongly associated with CRP levels (% difference per minor allele): SNP rs6700896 in LEPR (-14.8%; 95% confidence interval [CI], -17.6% to -12.0%; P = 6.2 x 10(-22)), rs4537545 in IL6R (-11.5%; 95% CI, -14.4% to -8.5%; P = 1.3 x 10(-12)), rs7553007 in the CRP locus (-20.7%; 95% CI, -23.4% to -17.9%; P = 1.3 x 10(-38)), rs1183910 in HNF1A (-13.8%; 95% CI, -16.6% to -10.9%; P = 1.9 x 10(-18)), and rs4420638 in APOE-CI-CII (-21.8%; 95% CI, -25.3% to -18.1%; P = 8.1 x 10(-26)). Association of SNP rs7553007 in the CRP locus with coronary heart disease gave an odds ratio (OR) of 0.98 (95% CI, 0.94 to 1.01) per 20% lower CRP level. Our mendelian randomization study of variants in the CRP locus showed no association with coronary heart disease: OR, 1.00; 95% CI, 0.97 to 1.02; per 20% lower CRP level, compared with OR, 0.94; 95% CI, 0.94 to 0.95; predicted from meta-analysis of the observational studies of CRP levels and coronary heart disease (z score, -3.45; P < .001). SNPs rs6700896 in LEPR (OR, 1.06; 95% CI, 1.02 to 1.09; per minor allele), rs4537545 in IL6R (OR, 0.94; 95% CI, 0.91 to 0.97), and rs4420638 in the APOE-CI-CII cluster (OR, 1.16; 95% CI, 1.12 to 1.21) were all associated with risk of coronary heart disease.
Conclusion: The lack of concordance between the effect on coronary heart disease risk of CRP genotypes and CRP levels argues against a causal association of CRP with coronary heart disease.
Figures
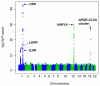
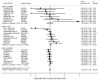




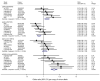
Comment in
-
Biomarkers and cardiovascular disease: determining causality and quantifying contribution to risk assessment.JAMA. 2009 Jul 1;302(1):92-3. doi: 10.1001/jama.2009.949. JAMA. 2009. PMID: 19567447 No abstract available.
References
-
- Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet. 1997;349(9061):1269–76. - PubMed
-
- Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352(16):1685–95. - PubMed
-
- Verma S, Szmitko PE, Ridker PM. C-reactive protein comes of age. Nat Clin Pract Cardiovasc Med. 2005;2(1):29–36. - PubMed
-
- Danesh J, Wheeler JG, Hirschfield GM, et al. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350(14):1387–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- G0600331/MRC_/Medical Research Council/United Kingdom
- R01 HL087679/HL/NHLBI NIH HHS/United States
- SP/04/002/BHF_/British Heart Foundation/United Kingdom
- G0700931/MRC_/Medical Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
- 1RL1MH083268-01/MH/NIMH NIH HHS/United States
- 089061/WT_/Wellcome Trust/United Kingdom
- CRUK_/Cancer Research UK/United Kingdom
- LSHM33 CT-2007-037273/BHF_/British Heart Foundation/United Kingdom
- G0801056/MRC_/Medical Research Council/United Kingdom
- MC_U137686857/MRC_/Medical Research Council/United Kingdom
- G0601966/MRC_/Medical Research Council/United Kingdom
- 5R01HL087679-02/HL/NHLBI NIH HHS/United States
- RL1 MH083268/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

