The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation
- PMID: 19567705
- PMCID: PMC2714929
- DOI: 10.1105/tpc.109.067785
The PLASTID DIVISION1 and 2 components of the chloroplast division machinery determine the rate of chloroplast division in land plant cell differentiation
Abstract
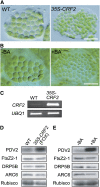
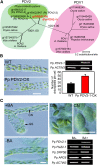
In most algae, the chloroplast division rate is held constant to maintain the proper number of chloroplasts per cell. By contrast, land plants evolved cell and chloroplast differentiation systems in which the size and number of chloroplasts change along with their respective cellular function by regulation of the division rate. Here, we show that PLASTID DIVISION (PDV) proteins, land plant-specific components of the division apparatus, determine the rate of chloroplast division. Overexpression of PDV proteins in the angiosperm Arabidopsis thaliana and the moss Physcomitrella patens increased the number but decreased the size of chloroplasts; reduction of PDV levels resulted in the opposite effect. The level of PDV proteins, but not other division components, decreased during leaf development, during which the chloroplast division rate also decreased. Exogenous cytokinins or overexpression of the cytokinin-responsive transcription factor CYTOKININ RESPONSE FACTOR2 increased the chloroplast division rate, where PDV proteins, but not other components of the division apparatus, were upregulated. These results suggest that the integration of PDV proteins into the division machinery enabled land plant cells to change chloroplast size and number in accord with the fate of cell differentiation.
Figures





Comment in
-
The evolution of the regulatory mechanism of chloroplast division.Plant Signal Behav. 2010 Feb;5(2):164-7. doi: 10.4161/psb.5.2.10461. Epub 2010 Feb 28. Plant Signal Behav. 2010. PMID: 20023413 Free PMC article.
References
-
- Boffey, S.A., and Lloyd, D. (1988). Division and Segregation of Organelles. (Cambridge, UK: Cambridge University Press).
-
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. - PubMed
-
- Colletti, K.S., Tattersall, E.A., Pyke, K.A., Froelich, J.E., Stokes, K.D., and Osteryoung, K.W. (2000). A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr. Biol. 10 507–516. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

