Epigenetic regulation of the alternatively activated macrophage phenotype
- PMID: 19567879
- PMCID: PMC2759649
- DOI: 10.1182/blood-2009-04-217620
Epigenetic regulation of the alternatively activated macrophage phenotype
Abstract
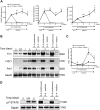
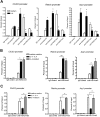
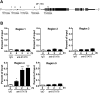
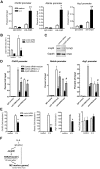
Alternatively activated (M2) macrophages play critical roles in diverse chronic diseases, including parasite infections, cancer, and allergic responses. However, little is known about the acquisition and maintenance of their phenotype. We report that M2-macrophage marker genes are epigenetically regulated by reciprocal changes in histone H3 lysine-4 (H3K4) and histone H3 lysine-27 (H3K27) methylation; and the latter methylation marks are removed by the H3K27 demethylase Jumonji domain containing 3 (Jmjd3). We found that continuous interleukin-4 (IL-4) treatment leads to decreased H3K27 methylation, at the promoter of M2 marker genes, and a concomitant increase in Jmjd3 expression. Furthermore, we demonstrate that IL-4-dependent Jmjd3 expression is mediated by STAT6, a major transcription factor of IL-4-mediated signaling. After IL-4 stimulation, activated STAT6 is increased and binds to consensus sites at the Jmjd3 promoter. Increased Jmjd3 contributes to the decrease of H3K27 dimethylation and trimethylation (H3K27me2/3) marks as well as the transcriptional activation of specific M2 marker genes. The decrease in H3K27me2/3 and increase in Jmjd3 recruitment were confirmed by in vivo studies using a Schistosoma mansoni egg-challenged mouse model, a well-studied system known to support an M2 phenotype. Collectively, these data indicate that chromatin remodeling is mechanistically important in the acquisition of the M2-macrophage phenotype.
Figures



Comment in
-
Orchestration of macrophage polarization.Blood. 2009 Oct 8;114(15):3135-6. doi: 10.1182/blood-2009-07-231795. Blood. 2009. PMID: 19815678
References
-
- Ramalingam TR, Reiman RM, Wynn TA. Exploiting worm and allergy models to understand Th2 cytokine biology. Curr Opin Allergy Clin Immunol. 2005;5:392–398. - PubMed
-
- Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. - PubMed
-
- Mantovani A, Sica A, Locati M. New vistas on macrophage differentiation and activation. Eur J Immunol. 2007;37:14–16. - PubMed
-
- Noel W, Raes G, Hassanzadeh Ghassabeh G, De Baetselier P, Beschin A. Alternatively activated macrophages during parasite infections. Trends Parasitol. 2004;20:126–133. - PubMed
-
- Zhu Z, Zheng T, Homer RJ, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

