Measuring and interpreting the selectivity of protein kinase inhibitors
- PMID: 19568781
- PMCID: PMC2725273
- DOI: 10.1007/s12154-009-0023-9
Measuring and interpreting the selectivity of protein kinase inhibitors
Abstract
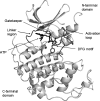
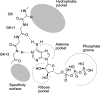
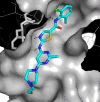
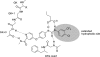
Protein kinase inhibitors are a well-established class of clinically useful drugs, particularly for the treatment of cancer. Achieving inhibitor selectivity for particular protein kinases often remains a significant challenge in the development of new small molecules as drugs or as tools for chemical biology research. This review summarises the methodologies available for measuring kinase inhibitor selectivity, both in vitro and in cells. The interpretation of kinase inhibitor selectivity data is discussed, particularly with reference to the structural biology of the protein targets. Measurement and prediction of kinase inhibitor selectivity will be important for the development of new multi-targeted kinase inhibitors.
Figures







References
LinkOut - more resources
Full Text Sources
Other Literature Sources

