Epigenetic abnormalities in cardiac hypertrophy and heart failure
- PMID: 19568876
- PMCID: PMC2698246
- DOI: 10.1007/s12199-007-0007-8
Epigenetic abnormalities in cardiac hypertrophy and heart failure
Abstract
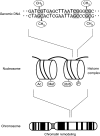
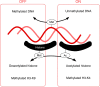
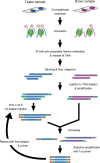
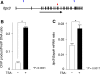
Epigenetics refers to the heritable regulation of gene expression through modification of chromosomal components without an alteration in the nucleotide sequence of the genome. Such modifications include methylation of genomic DNA as well as acetylation, methylation, phosphorylation, ubiquitination, and SUMOylation of core histone proteins. Recent genetic and biochemical analyses indicate that epigenetic changes play an important role in the development of cardiac hypertrophy and heart failure, with dysregulation in histone acetylation status, in particular, shown to be directly linked to an impaired contraction ability of cardiac myocytes. Although such epigenetic changes should eventually lead to alterations in the expression of genes associated with the affected histones, little information is yet available on the genes responsible for the development of heart failure. Current efforts of our and other groups have focused on deciphering the network of genes which are under abnormal epigenetic regulation in failed hearts. To this end, coupling chromatin immunoprecipitation to high-throughput profiling systems is being applied to cardiac myocytes in normal as well as affected hearts. The results of these studies should not only improve our understanding of the molecular basis for cardiac hypertrophy/heart failure but also provide essential information that will facilitate the development of new epigenetics-based therapies.
Figures

References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1038/nature02625', 'is_inner': False, 'url': 'https://doi.org/10.1038/nature02625'}, {'type': 'PubMed', 'value': '15164071', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15164071/'}]}
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1038/47412', 'is_inner': False, 'url': 'https://doi.org/10.1038/47412'}, {'type': 'PubMed', 'value': '10638745', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/10638745/'}]}
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–5. - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1073/pnas.0500136102', 'is_inner': False, 'url': 'https://doi.org/10.1073/pnas.0500136102'}, {'type': 'PMC', 'value': 'PMC555684', 'is_inner': False, 'url': 'https://pmc.ncbi.nlm.nih.gov/articles/PMC555684/'}, {'type': 'PubMed', 'value': '15795371', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15795371/'}]}
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci USA. 2005;102:5501–6. - PMC - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1016/S0168-9525(03)00073-8', 'is_inner': False, 'url': 'https://doi.org/10.1016/s0168-9525(03)00073-8'}, {'type': 'PubMed', 'value': '12711221', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12711221/'}]}
- Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–93. - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1139/o02-080', 'is_inner': False, 'url': 'https://doi.org/10.1139/o02-080'}, {'type': 'PubMed', 'value': '12123289', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/12123289/'}]}
- Waterborg JH. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem Cell Biol. 2002;80:363–78. - PubMed
