Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli
- PMID: 19570135
- PMCID: PMC2770234
- DOI: 10.1111/j.1365-2958.2009.06781.x
Hfq affects the expression of the LEE pathogenicity island in enterohaemorrhagic Escherichia coli
Abstract
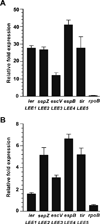
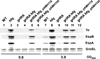
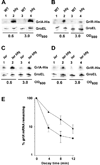
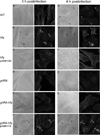
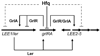
Colonization of the intestinal epithelium by enterohaemorrhagic Escherichia coli (EHEC) is characterized by an attaching and effacing (A/E) histopathology. The locus of enterocyte effacement (LEE) pathogenicity island encodes many genes required for the A/E phenotype including the global regulator of EHEC virulence gene expression, Ler. The LEE is subject to a complex regulatory network primarily targeting ler transcription. The RNA chaperone Hfq, implicated in post-transcriptional regulation, is an important virulence factor in many bacterial pathogens. Although post-transcriptional regulation of EHEC virulence genes is known to occur, a regulatory role of Hfq in EHEC virulence gene expression has yet to be defined. Here, we show that an hfq mutant expresses increased levels of LEE-encoded proteins prematurely, leading to earlier A/E lesion formation relative to wild type. Hfq indirectly affects LEE expression in exponential phase independent of Ler by negatively controlling levels of the regulators GrlA and GrlR through post-transcriptional regulation of the grlRA messenger. Moreover, Hfq negatively affects LEE expression in stationary phase independent of GrlA and GrlR. Altogether, Hfq plays an important role in co-ordinating the temporal expression of the LEE by controlling grlRA expression at the post-transcriptional level.
Figures



Similar articles
-
RNA-binding protein Hfq downregulates locus of enterocyte effacement-encoded regulators independent of small regulatory RNA in enterohemorrhagic Escherichia coli.Mol Microbiol. 2022 Jan;117(1):86-101. doi: 10.1111/mmi.14799. Epub 2021 Aug 31. Mol Microbiol. 2022. PMID: 34411346
-
Hfq negatively regulates type III secretion in EHEC and several other pathogens.Mol Microbiol. 2009 Oct;74(2):347-63. doi: 10.1111/j.1365-2958.2009.06856.x. Epub 2009 Aug 24. Mol Microbiol. 2009. PMID: 19703108 Free PMC article.
-
Coordinate control of the locus of enterocyte effacement and enterohemolysin genes by multiple common virulence regulators in enterohemorrhagic Escherichia coli.Infect Immun. 2011 Nov;79(11):4628-37. doi: 10.1128/IAI.05023-11. Epub 2011 Aug 15. Infect Immun. 2011. PMID: 21844237 Free PMC article.
-
The Tip of the Iceberg: On the Roles of Regulatory Small RNAs in the Virulence of Enterohemorrhagic and Enteropathogenic Escherichia coli.Front Cell Infect Microbiol. 2016 Sep 21;6:105. doi: 10.3389/fcimb.2016.00105. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27709103 Free PMC article. Review.
-
Control freaks-signals and cues governing the regulation of virulence in attaching and effacing pathogens.Biochem Soc Trans. 2019 Feb 28;47(1):229-238. doi: 10.1042/BST20180546. Epub 2018 Dec 17. Biochem Soc Trans. 2019. PMID: 30559275 Free PMC article. Review.
Cited by
-
Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli.J Biol Chem. 2018 Jun 8;293(23):9006-9016. doi: 10.1074/jbc.RA117.000300. Epub 2018 Apr 20. J Biol Chem. 2018. PMID: 29678883 Free PMC article.
-
RpoS role in virulence and fitness in enteropathogenic Escherichia coli.PLoS One. 2017 Jun 29;12(6):e0180381. doi: 10.1371/journal.pone.0180381. eCollection 2017. PLoS One. 2017. PMID: 28662183 Free PMC article.
-
An RNA-dependent mechanism for transient expression of bacterial translocation filaments.Nucleic Acids Res. 2018 Apr 20;46(7):3366-3381. doi: 10.1093/nar/gky096. Nucleic Acids Res. 2018. PMID: 29432565 Free PMC article.
-
Determining the relative contribution and hierarchy of hha and qseBC in the regulation of flagellar motility of Escherichia coli O157:H7.PLoS One. 2014 Jan 21;9(1):e85866. doi: 10.1371/journal.pone.0085866. eCollection 2014. PLoS One. 2014. PMID: 24465756 Free PMC article.
-
The E. coli transcription factor GrlA is regulated by subcellular compartmentalization and activated in response to mechanical stimuli.Proc Natl Acad Sci U S A. 2020 Apr 28;117(17):9519-9528. doi: 10.1073/pnas.1917500117. Epub 2020 Apr 10. Proc Natl Acad Sci U S A. 2020. PMID: 32277032 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

