Prognostic relevance of Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter methylation in human breast cancer
- PMID: 19570204
- PMCID: PMC2717119
- DOI: 10.1186/1471-2407-9-217
Prognostic relevance of Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter methylation in human breast cancer
Abstract
Background: Secreted Wnt signaling antagonists have recently been described as frequent targets of epigenetic inactivation in human tumor entities. Since gene silencing of certain Wnt antagonists was found to be correlated with adverse patient survival in cancer, we aimed at investigating a potential prognostic impact of the two Wnt antagonizing molecules WIF1 and DKK3 in breast cancer, which are frequently silenced by promoter methylation in this disease.
Methods: WIF1 and DKK3 promoter methylation were assessed by methylation-specific PCR with bisulfite-converted DNA from 19 normal breast tissues and 150 primary breast carcinomas. Promoter methylation was interpreted in a qualitative, binary fashion. Statistical evaluations included two-sided Fisher's exact tests, univariate log-rank tests of Kaplan-Meier curves as well as multivariate Cox regression analyses.
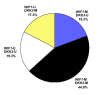
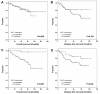
Results: WIF1 and DKK3 promoter methylation were detected in 63.3% (95/150) and 61.3% (92/150) of breast carcinoma samples, respectively. In normal breast tissues, WIF1 methylation was present in 0% (0/19) and DKK3 methylation in 5.3% (1/19) of samples. In breast carcinomas, WIF1 methylation was significantly associated with methylation of DKK3 (p = 0.009). Methylation of either gene was not associated with clinicopathological parameters, except for DKK3 methylation being associated with patient age (p = 0.007). In univariate analysis, WIF1 methylation was not associated with clinical patient outcome. In contrast, DKK3 methylation was a prognostic factor in patient overall survival (OS) and disease-free survival (DFS). Estimated OS rates after 10 years were 54% for patients with DKK3-methylated tumors, in contrast to patients without DKK3 methylation in the tumor, who had a favorable 97% OS after 10 years (p < 0.001). Likewise, DFS at 10 years for patients harboring DKK3 methylation in the tumor was 58%, compared with 78% for patients with unmethylated DKK3 (p = 0.037). Multivariate analyses revealed that DKK3 methylation was an independent prognostic factor predicting poor OS (hazard ratio (HR): 14.4; 95% confidence interval (CI): 1.9-111.6; p = 0.011), and short DFS (HR: 2.5; 95% CI: 1.0-6.0; p = 0.047) in breast cancer.
Conclusion: Although the Wnt antagonist genes WIF1 and DKK3 show a very similar frequency of promoter methylation in human breast cancer, only DKK3 methylation proves as a novel prognostic marker potentially useful in the clinical management of this disease.
Figures



References
-
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. - PubMed
-
- Howe LR, Brown AM. Wnt signaling and breast cancer. Cancer Biol Ther. 2004;3:36–41. - PubMed
-
- Aguilera O, Muñoz A, Esteller M, Fraga MF. Epigenetic alterations of the Wnt/beta-catenin pathway in human disease. Endocr Metab Immune Disord Drug Targets. 2007;7:13–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

