Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model
- PMID: 19570746
- PMCID: PMC2726834
- DOI: 10.1093/molbev/msp123
Streamlining and large ancestral genomes in Archaea inferred with a phylogenetic birth-and-death model
Abstract
Homologous genes originate from a common ancestor through vertical inheritance, duplication, or horizontal gene transfer. Entire homolog families spawned by a single ancestral gene can be identified across multiple genomes based on protein sequence similarity. The sequences, however, do not always reveal conclusively the history of large families. To study the evolution of complete gene repertoires, we propose here a mathematical framework that does not rely on resolved gene family histories. We show that so-called phylogenetic profiles, formed by family sizes across multiple genomes, are sufficient to infer principal evolutionary trends. The main novelty in our approach is an efficient algorithm to compute the likelihood of a phylogenetic profile in a model of birth-and-death processes acting on a phylogeny. We examine known gene families in 28 archaeal genomes using a probabilistic model that involves lineage- and family-specific components of gene acquisition, duplication, and loss. The model enables us to consider all possible histories when inferring statistics about archaeal evolution. According to our reconstruction, most lineages are characterized by a net loss of gene families. Major increases in gene repertoire have occurred only a few times. Our reconstruction underlines the importance of persistent streamlining processes in shaping genome composition in Archaea. It also suggests that early archaeal genomes were as complex as typical modern ones, and even show signs, in the case of the methanogenic ancestor, of an extremely large gene repertoire.
Figures

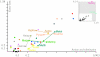
 compared with expected numbers of substitutions (or edge length) for each branch e. Pairs of sibling terminal taxa are connected by lines.
compared with expected numbers of substitutions (or edge length) for each branch e. Pairs of sibling terminal taxa are connected by lines.
References
-
- Alexeyenko A, Tamas I, Liu G, Sonnhammer ELL. Automatic clustering of orthologs and inparalogs shared by multiple genomes. Bioinformatics. 2006;22:e9–e15. - PubMed
-
- Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau MER, Nesbo CL, Case RJ, Doolittle WF. Lateral gene transfer and the origin of prokaryotic groups. Ann Rev Genet. 2003;37:283–328. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

