Functional analysis of N-terminal residues of ty1 integrase
- PMID: 19570857
- PMCID: PMC2738233
- DOI: 10.1128/JVI.00159-09
Functional analysis of N-terminal residues of ty1 integrase
Abstract
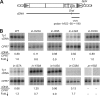
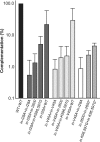
The Ty1 retrotransposon of Saccharomyces cerevisiae is comprised of structural and enzymatic proteins that are functionally similar to those of retroviruses. Despite overall sequence divergence, certain motifs are highly conserved. We have examined the Ty1 integrase (IN) zinc binding domain by mutating the definitive histidine and cysteine residues and thirteen residues in the intervening (X(32)) sequence between IN-H22 and IN-C55. Mutation of the zinc-coordinating histidine or cysteine residues reduced transposition by more than 4,000-fold and led to IN and reverse transcriptase (RT) instability as well as inefficient proteolytic processing. Alanine substitution of the hydrophobic residues I28, L32, I37 and V45 in the X(32) region reduced transposition 85- to 688-fold. Three of these residues, L32, I37, and V45, are highly conserved among retroviruses, although their effects on integration or viral infectivity have not been characterized. In contrast to the HHCC mutants, all the X(32) mutants exhibited stable IN and RT, and protein processing and cDNA production were unaffected. However, glutathione S-transferase pulldowns and intragenic complementation analysis of selected transposition-defective X(32) mutants revealed decreased IN-IN interactions. Furthermore, virus-like particles with in-L32A and in-V45A mutations did not exhibit substantial levels of concerted integration products in vitro. Our results suggest that the histidine/cysteine residues are important for steps in transposition prior to integration, while the hydrophobic residues function in IN multimerization.
Figures




Similar articles
-
Ty1 integrase is composed of an active N-terminal domain and a large disordered C-terminal module dispensable for its activity in vitro.J Biol Chem. 2021 Oct;297(4):101093. doi: 10.1016/j.jbc.2021.101093. Epub 2021 Aug 17. J Biol Chem. 2021. PMID: 34416236 Free PMC article.
-
Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo.Mol Cell Biol. 1998 Feb;18(2):1115-24. doi: 10.1128/MCB.18.2.1115. Mol Cell Biol. 1998. PMID: 9448009 Free PMC article.
-
A Ty1 integrase nuclear localization signal required for retrotransposition.Mol Cell Biol. 1998 Feb;18(2):1105-14. doi: 10.1128/MCB.18.2.1105. Mol Cell Biol. 1998. PMID: 9448008 Free PMC article.
-
Reverse transcriptase and integrase of the Saccharomyces cerevisiae Ty1 element.Cytogenet Genome Res. 2005;110(1-4):269-87. doi: 10.1159/000084960. Cytogenet Genome Res. 2005. PMID: 16093680 Review.
-
Fidelity of retrotransposon replication.Ann N Y Acad Sci. 1999 May 18;870:108-18. doi: 10.1111/j.1749-6632.1999.tb08871.x. Ann N Y Acad Sci. 1999. PMID: 10415477 Review.
Cited by
-
Ty1 integrase is composed of an active N-terminal domain and a large disordered C-terminal module dispensable for its activity in vitro.J Biol Chem. 2021 Oct;297(4):101093. doi: 10.1016/j.jbc.2021.101093. Epub 2021 Aug 17. J Biol Chem. 2021. PMID: 34416236 Free PMC article.
-
The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae.Microbiol Spectr. 2015 Apr 1;3(2):1-35. doi: 10.1128/microbiolspec.MDNA3-0053-2014. Microbiol Spectr. 2015. PMID: 25893143 Free PMC article.
-
BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae.Genetics. 2010 Aug;185(4):1193-205. doi: 10.1534/genetics.110.119115. Epub 2010 May 24. Genetics. 2010. PMID: 20498295 Free PMC article.
-
The Pu.1 target gene Zbtb11 regulates neutrophil development through its integrase-like HHCC zinc finger.Nat Commun. 2017 Apr 6;8:14911. doi: 10.1038/ncomms14911. Nat Commun. 2017. PMID: 28382966 Free PMC article.
References
-
- Adams, S. E., J. Mellor, K. Gull, R. B. Sim, M. F. Tuite, S. M. Kingsman, and A. J. Kingsman. 1987. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 49:111-119. - PubMed
-
- Andrake, M. D., and A. M. Skalka. 1995. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J. Biol. Chem. 270:29299-29306. - PubMed
-
- Bertola, F., C. Manigand, P. Picard, M. Goetz, J.-M. Schmitter, and G. Precigoux. 2001. N-terminal domain of HTLV-1 integrase. Complexation and conformational studies of the zinc finger. J. Peptide Sci. 7:588-597. - PubMed
-
- Bizub-Bender, D., J. Kulkosky, and A. M. Skalka. 1994. Monoclonal antibodies against HIV type 1 integrase: clues to molecular structure. AIDS Res. Hum. Retroviruses 10:1105-1115. - PubMed
-
- Boeke, J. D., D. J. Garfinkel, C. A. Styles, and G. R. Fink. 1985. Ty elements transpose through an RNA intermediate. Cell 40:491-500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

