Role of oxysterol binding protein in hepatitis C virus infection
- PMID: 19570870
- PMCID: PMC2738263
- DOI: 10.1128/JVI.00958-09
Role of oxysterol binding protein in hepatitis C virus infection
Abstract
Hepatitis C virus (HCV) RNA genome replicates within the ribonucleoprotein (RNP) complex in the modified membranous structures extended from endoplasmic reticulum. A proteomic analysis of HCV RNP complexes revealed the association of oxysterol binding protein (OSBP) as one of the components of these complexes. OSBP interacted with the N-terminal domain I of the HCV NS5A protein and colocalized to the Golgi compartment with NS5A. An OSBP-specific short hairpin RNA that partially downregulated OSBP expression resulted in a decrease of the HCV particle release in culture supernatant with little effect on viral RNA replication. The pleckstrin homology (PH) domain located in the N-terminal region of OSBP targeted this protein to the Golgi apparatus. OSBP deletion mutation in the PH (DeltaPH) domain failed to localize to the Golgi apparatus and inhibited the HCV particle release. These studies suggest a possible functional role of OSBP in the HCV maturation process.
Figures
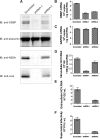

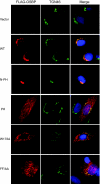


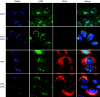
Similar articles
-
Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking.Gastroenterology. 2014 May;146(5):1373-85.e1-11. doi: 10.1053/j.gastro.2014.02.002. Epub 2014 Feb 8. Gastroenterology. 2014. PMID: 24512803 Free PMC article.
-
Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein.J Biol Chem. 2011 Apr 1;286(13):11265-74. doi: 10.1074/jbc.M110.182097. Epub 2011 Feb 1. J Biol Chem. 2011. PMID: 21285358 Free PMC article.
-
Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum.J Biol Chem. 2002 Aug 16;277(33):29908-18. doi: 10.1074/jbc.M201191200. Epub 2002 May 21. J Biol Chem. 2002. PMID: 12023275
-
Phosphoinositides in the hepatitis C virus life cycle.Viruses. 2012 Oct 19;4(10):2340-58. doi: 10.3390/v4102340. Viruses. 2012. PMID: 23202467 Free PMC article. Review.
-
Bridging the molecular and biological functions of the oxysterol-binding protein family.Cell Mol Life Sci. 2018 Sep;75(17):3079-3098. doi: 10.1007/s00018-018-2795-y. Epub 2018 Mar 13. Cell Mol Life Sci. 2018. PMID: 29536114 Free PMC article. Review.
Cited by
-
Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion.J Biol Chem. 2012 Aug 10;287(33):27637-47. doi: 10.1074/jbc.M112.346569. Epub 2012 Jun 28. J Biol Chem. 2012. PMID: 22745132 Free PMC article.
-
Evaluation of canonical siRNA and Dicer substrate RNA for inhibition of hepatitis C virus genome replication--a comparative study.PLoS One. 2015 Feb 23;10(2):e0117742. doi: 10.1371/journal.pone.0117742. eCollection 2015. PLoS One. 2015. PMID: 25705875 Free PMC article.
-
MEAN inhibits hepatitis C virus replication by interfering with a polypyrimidine tract-binding protein.J Cell Mol Med. 2016 Jul;20(7):1255-65. doi: 10.1111/jcmm.12798. Epub 2016 Mar 1. J Cell Mol Med. 2016. PMID: 26929148 Free PMC article.
-
Transient Compound Treatment Induces a Multigenerational Reduction of Oxysterol-Binding Protein (OSBP) Levels and Prophylactic Antiviral Activity.ACS Chem Biol. 2019 Feb 15;14(2):276-287. doi: 10.1021/acschembio.8b00984. Epub 2019 Jan 11. ACS Chem Biol. 2019. PMID: 30576108 Free PMC article.
-
Evaluation of ITX 5061, a scavenger receptor B1 antagonist: resistance selection and activity in combination with other hepatitis C virus antivirals.J Infect Dis. 2012 Feb 15;205(4):656-62. doi: 10.1093/infdis/jir802. J Infect Dis. 2012. PMID: 22279172 Free PMC article.
References
-
- Aizaki, H., K. Morikawa, M. Fukasawa, H. Hara, Y. Inoue, H. Tani, K. Saito, M. Nishijima, K. Hanada, Y. Matsuura, M. M. Lai, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J. Virol. 82:5715-5724. - PMC - PubMed
-
- Bern, M., D. Goldberg, W. H. McDonald, and J. R. Yates, 3rd. 2004. Automatic quality assessment of peptide tandem mass spectra. Bioinformatics 20(Suppl. 1):i49-i54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

