Subcellular localization of the barley stripe mosaic virus triple gene block proteins
- PMID: 19570874
- PMCID: PMC2738231
- DOI: 10.1128/JVI.00739-09
Subcellular localization of the barley stripe mosaic virus triple gene block proteins
Erratum in
- J Virol. 2009 Nov;83(21):11413
Abstract
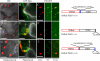
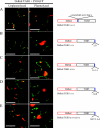
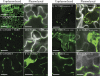
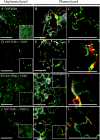
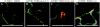
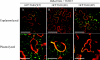
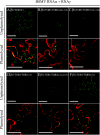
Barley stripe mosaic virus (BSMV) spreads from cell to cell through the coordinated actions of three triple gene block (TGB) proteins (TGB1, TGB2, and TGB3) arranged in overlapping open reading frames (ORFs). Our previous studies (D. M. Lawrence and A. O. Jackson, J. Virol. 75:8712-8723, 2001; D. M. Lawrence and A. O. Jackson, Mol. Plant Pathol. 2:65-75, 2001) have shown that each of these proteins is required for cell-to-cell movement in monocot and dicot hosts. We recently found (H.-S. Lim, J. N. Bragg, U. Ganesan, D. M. Lawrence, J. Yu, M. Isogai, J. Hammond, and A. O. Jackson, J. Virol. 82:4991-5006, 2008) that TGB1 engages in homologous interactions leading to the formation of a ribonucleoprotein complex containing viral genomic and messenger RNAs, and we have also demonstrated that TGB3 functions in heterologous interactions with TGB1 and TGB2. We have now used Agrobacterium tumefaciens-mediated protein expression in Nicotiana benthamiana leaf cells and site-specific mutagenesis to determine how TGB protein interactions influence their subcellular localization and virus spread. Confocal microscopy revealed that the TGB3 protein localizes at the cell wall (CW) in close association with plasmodesmata and that the deletion or mutagenesis of a single amino acid at the immediate C terminus can affect CW targeting. TGB3 also directed the localization of TGB2 from the endoplasmic reticulum to the CW, and this targeting was shown to be dependent on interactions between the TGB2 and TGB3 proteins. The optimal localization of the TGB1 protein at the CW also required TGB2 and TGB3 interactions, but in this context, site-specific TGB1 helicase motif mutants varied in their localization patterns. The results suggest that the ability of TGB1 to engage in homologous binding interactions is not essential for targeting to the CW. However, the relative expression levels of TGB2 and TGB3 influenced the cytosolic and CW distributions of TGB1 and TGB2. Moreover, in both cases, localization at the CW was optimal at the 10:1 TGB2-to-TGB3 ratios occurring in virus infections, and mutations reducing CW localization had corresponding effects on BSMV movement phenotypes. These data support a model whereby TGB protein interactions function in the subcellular targeting of movement protein complexes and the ability of BSMV to move from cell to cell.
Figures



References
-
- Baulcombe, D. C. 2004. RNA silencing in plants. Nature 431:356-363. - PubMed
-
- Bleykasten, C., D. Gilmer, H. Guilley, K. E. Richards, and G. Jonard. 1996. Beet necrotic yellow vein virus 42 kDa triple gene block protein binds nucleic acid in vitro. J. Gen. Virol. 77:889-897. - PubMed
-
- Bragg, J. N., and A. O. Jackson. 2004. The C-terminal region of the Barley stripe mosaic virus γb protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5:465-481. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

