Distinct roles for key karyogamy proteins during yeast nuclear fusion
- PMID: 19570912
- PMCID: PMC2735476
- DOI: 10.1091/mbc.e09-02-0163
Distinct roles for key karyogamy proteins during yeast nuclear fusion
Abstract
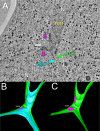
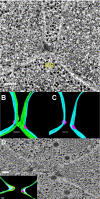
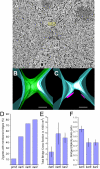
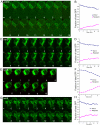
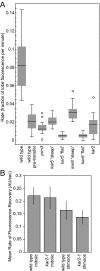
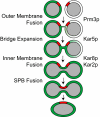
During yeast mating, cell fusion is followed by the congression and fusion of the two nuclei. Proteins required for nuclear fusion are found at the surface (Prm3p) and within the lumen (Kar2p, Kar5p, and Kar8p) of the nuclear envelope (NE). Electron tomography (ET) of zygotes revealed that mutations in these proteins block nuclear fusion with different morphologies, suggesting that they act in different steps of fusion. Specifically, prm3 zygotes were blocked before formation of membrane bridges, whereas kar2, kar5, and kar8 zygotes frequently contained them. Membrane bridges were significantly larger and occurred more frequently in kar2 and kar8, than in kar5 mutant zygotes. The kinetics of NE fusion in prm3, kar5, and kar8 mutants, measured by live-cell fluorescence microscopy, were well correlated with the size and frequency of bridges observed by ET. However the kar2 mutant was defective for transfer of NE lumenal GFP, but not diffusion within the lumen, suggesting that transfer was blocked at the NE fusion junction. These observations suggest that Prm3p acts before initiation of outer NE fusion, Kar5p may help dilation of the initial fusion pore, and Kar2p and Kar8p act after outer NE fusion, during inner NE fusion.
Figures


Similar articles
-
Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae.Mol Biol Cell. 1999 Mar;10(3):609-26. doi: 10.1091/mbc.10.3.609. Mol Biol Cell. 1999. PMID: 10069807 Free PMC article.
-
Kar5p is required for multiple functions in both inner and outer nuclear envelope fusion in Saccharomyces cerevisiae.G3 (Bethesda). 2014 Dec 2;5(1):111-21. doi: 10.1534/g3.114.015800. G3 (Bethesda). 2014. PMID: 25467943 Free PMC article.
-
Prm3p is a pheromone-induced peripheral nuclear envelope protein required for yeast nuclear fusion.Mol Biol Cell. 2009 May;20(9):2438-50. doi: 10.1091/mbc.e08-10-0987. Epub 2009 Mar 18. Mol Biol Cell. 2009. PMID: 19297527 Free PMC article.
-
Nuclear fusion in the yeast Saccharomyces cerevisiae.Annu Rev Cell Dev Biol. 1996;12:663-95. doi: 10.1146/annurev.cellbio.12.1.663. Annu Rev Cell Dev Biol. 1996. PMID: 8970740 Review.
-
The Malleable Nature of the Budding Yeast Nuclear Envelope: Flares, Fusion, and Fenestrations.J Cell Physiol. 2016 Nov;231(11):2353-60. doi: 10.1002/jcp.25355. Epub 2016 Apr 8. J Cell Physiol. 2016. PMID: 26909870 Free PMC article. Review.
Cited by
-
Cell biology of yeast zygotes, from genesis to budding.Biochim Biophys Acta. 2015 Jul;1853(7):1702-14. doi: 10.1016/j.bbamcr.2015.03.018. Epub 2015 Apr 8. Biochim Biophys Acta. 2015. PMID: 25862405 Free PMC article. Review.
-
Dynamics of Male and Female Chromatin during Karyogamy in Rice Zygotes.Plant Physiol. 2014 Aug;165(4):1533-1543. doi: 10.1104/pp.114.236059. Epub 2014 Jun 19. Plant Physiol. 2014. PMID: 24948834 Free PMC article.
-
When yeast cells meet, karyogamy!: an example of nuclear migration slowly resolved.Nucleus. 2013 May-Jun;4(3):182-8. doi: 10.4161/nucl.25021. Epub 2013 May 15. Nucleus. 2013. PMID: 23715006 Free PMC article. Review.
-
Nuclear Fusion in Yeast and Plant Reproduction.Plants (Basel). 2023 Oct 18;12(20):3608. doi: 10.3390/plants12203608. Plants (Basel). 2023. PMID: 37896071 Free PMC article.
-
Conservation of proteo-lipid nuclear membrane fusion machinery during early embryogenesis.Nucleus. 2014 Sep-Oct;5(5):441-8. doi: 10.4161/nucl.34422. Nucleus. 2014. PMID: 25482196 Free PMC article.
References
-
- Amberg D. C., Burke D. J., Strathern J. N. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005.
-
- Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:8219–8223. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

