The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods
- PMID: 19571240
- PMCID: PMC2873002
- DOI: 10.1098/rstb.2009.0034
The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods
Abstract
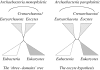
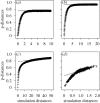
The three-domains tree, which depicts eukaryotes and archaebacteria as monophyletic sister groups, is the dominant model for early eukaryotic evolution. By contrast, the 'eocyte hypothesis', where eukaryotes are proposed to have originated from within the archaebacteria as sister to the Crenarchaeota (also called the eocytes), has been largely neglected in the literature. We have investigated support for these two competing hypotheses from molecular sequence data using methods that attempt to accommodate the across-site compositional heterogeneity and across-tree compositional and rate matrix heterogeneity that are manifest features of these data. When ribosomal RNA genes were analysed using standard methods that do not adequately model these kinds of heterogeneity, the three-domains tree was supported. However, this support was eroded or lost when composition-heterogeneous models were used, with concomitant increase in support for the eocyte tree for eukaryotic origins. Analysis of combined amino acid sequences from 41 protein-coding genes supported the eocyte tree, whether or not composition-heterogeneous models were used. The possible effects of substitutional saturation of our data were examined using simulation; these results suggested that saturation is delayed by among-site rate variation in the sequences, and that phylogenetic signal for ancient relationships is plausibly present in these data.
Figures




References
-
- Baldauf S. L., Palmer J. D., Doolittle W. F.1996The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl Acad. Sci. USA 93, 7749–7754 (doi:10.1073/pnas.93.15.7749) - DOI - PMC - PubMed
-
- Barns S. M., Delwiche C. F., Palmer J. D., Pace N. R.1996Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc. Natl Acad. Sci. USA 93, 9188–9193 (doi:10.1073/pnas.93.17.9188) - DOI - PMC - PubMed
-
- Bollback J. P.2002Bayesian model adequacy and choice in phylogenetics. Mol. Biol. Evol. 19, 1171–1180 - PubMed
-
- Brown J. R., Douady C. J., Italia M. J., Marshall W. E., Stanhope M. J.2001Universal trees based on large combined protein sequence data sets. Nat. Genet. 28, 281–285 (doi:10.1038/90129) - DOI - PubMed
-
- Cavalier-Smith T.2002The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

