Conjugative plasmids: vessels of the communal gene pool
- PMID: 19571247
- PMCID: PMC2873005
- DOI: 10.1098/rstb.2009.0037
Conjugative plasmids: vessels of the communal gene pool
Abstract
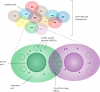
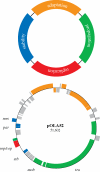
Comparative whole-genome analyses have demonstrated that horizontal gene transfer (HGT) provides a significant contribution to prokaryotic genome innovation. The evolution of specific prokaryotes is therefore tightly linked to the environment in which they live and the communal pool of genes available within that environment. Here we use the term supergenome to describe the set of all genes that a prokaryotic 'individual' can draw on within a particular environmental setting. Conjugative plasmids can be considered particularly successful entities within the communal pool, which have enabled HGT over large taxonomic distances. These plasmids are collections of discrete regions of genes that function as 'backbone modules' to undertake different aspects of overall plasmid maintenance and propagation. Conjugative plasmids often carry suites of 'accessory elements' that contribute adaptive traits to the hosts and, potentially, other resident prokaryotes within specific environmental niches. Insight into the evolution of plasmid modules therefore contributes to our knowledge of gene dissemination and evolution within prokaryotic communities. This communal pool provides the prokaryotes with an important mechanistic framework for obtaining adaptability and functional diversity that alleviates the need for large genomes of specialized 'private genes'.
Figures

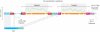
References
-
- Allen E. E., Tyson G. W., Whitaker R. J., Detter J. C., Richardson P. M., Banfield J. F.2007Genome dynamics in a natural archaeal population. Proc. Natl Acad. Sci. 104, 1883–1888 (doi:10.1073/pnas.0604851104) - DOI - PMC - PubMed
-
- Alonso G., Baptista K., Ngo T., Taylor D. E.2005Transcriptional organization of the temperature-sensitive transfer system from the IncHI1 plasmid R27. Microbiology 151, 3563–3573 (doi:10.1099/mic.0.28256-0) - DOI - PubMed
-
- Althorpe N. J., Chilley P. M., Thomas A. T., Brammar W. J., Wilkins B. M.1999Transient transcriptional activation of the Incl1 plasmid anti-restriction gene (ardA) and SOS inhibition gene (psiB) early in conjugating recipient bacteria. Mol. Microbiol. 31, 133–142 (doi:10.1046/j.1365-2958.1999.01153.x) - DOI - PubMed
-
- Bahl M. I., Hansen L. H., Licht T. R., Sørensen S. J.2007aConjugative transfer facilitates stable maintenance of IncP-1 plasmid pKJK5 in Escherichia coli cells colonizing the gastrointestinal tract of the germfree rat. Appl. Environ. Microbiol. 73, 341–343 (doi:10.1128/AEM.01971-06) - DOI - PMC - PubMed
-
- Bahl M. I., Hansen L. H., Sørensen S. J.2007bImpact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol. Lett. 266, 250–256 (doi:10.1111/j.1574-6968.2006.00536.x) - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
