Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean
- PMID: 19572557
- PMCID: PMC3086478
- DOI: 10.1021/ac900888s
Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean
Abstract
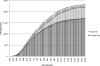
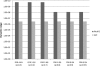
Data-dependent precursor ion selection is widely used in shotgun proteomics to profile the protein components of complex samples. Although very popular, this bottom-up method presents major drawbacks in terms of detectable dynamic range. Here, we demonstrate the superior performance of a data-independent method we term precursor acquisition independent from ion count (PAcIFIC). Our results show that almost the entire, predicted, soluble bacterial proteome can be thoroughly analyzed by PAcIFIC without the need for any sample fractionation other than the C18-based liquid chromatograph used to introduce the peptide mixture into the mass spectrometer. Importantly, we also show that PAcIFIC provides unique performance for analysis of human plasma in terms of the number of proteins identified (746 at FDR < or = 0.5%) and achieved dynamic range (8 orders of magnitude at FDR < or = 0.5%), without any fractionation other than immuno-depletion of the seven most abundant proteins.
Figures

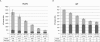
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

