Deoxyribozymes: selection design and serendipity in the development of DNA catalysts
- PMID: 19572701
- PMCID: PMC2764829
- DOI: 10.1021/ar900052y
Deoxyribozymes: selection design and serendipity in the development of DNA catalysts
Abstract
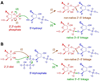
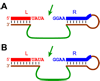
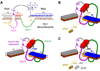
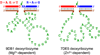
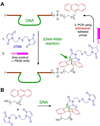
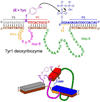
One of the chemist's key motivations is to explore the forefront of catalysis. In this Account, we describe our laboratory's efforts at one such forefront: the use of DNA as a catalyst. Natural biological catalysts include both protein enzymes and RNA enzymes (ribozymes), whereas nature apparently uses DNA solely for genetic information storage. Nevertheless, the chemical similarities between RNA and DNA naturally lead to laboratory examination of DNA as a catalyst, especially because DNA is more stable than RNA and is less costly and easier to synthesize. Many catalytically active DNA sequences (deoxyribozymes, also called DNAzymes) have been identified in the laboratory by in vitro selection, in which many random DNA sequences are evaluated in parallel to find those rare sequences that have a desired functional ability. Since 2001, our research group has pursued new deoxyribozymes for various chemical reactions. We consider DNA simply as a large biopolymer that can adopt intricate three-dimensional structure and, in the presence of appropriate metal ions, generate the chemical complexity required to achieve catalysis. Our initial efforts focused on deoxyribozymes that ligate two RNA substrates. In these studies, we used only substrates that are readily obtained biochemically. Highly active deoxyribozymes were identified, with emergent questions regarding chemical selectivity during RNA phosphodiester bond formation. Deoxyribozymes allow synthesis of interesting RNA products, such as branches and lariats, that are otherwise challenging to prepare. Our experiments have demonstrated that deoxyribozymes can have very high rate enhancements and chemical selectivities. We have also shown how the in vitro selection process itself can be directed toward desired goals, such as selective formation of native 3'-5' RNA linkages. A final lesson is that unanticipated selection outcomes can be very interesting, highlighting the importance of allowing such opportunities in future experiments. More recently, we have begun using nonoligonucleotide substrates in our efforts with deoxyribozymes. We have especially focused on developing DNA catalysts for reactions of small molecules or amino acid side chains. For example, new deoxyribozymes have the catalytic power to create a nucleopeptide linkage between a tyrosine or serine side chain and the 5'-terminus of an RNA strand. Although considerable further work remains to establish DNA as a practical catalyst for small molecules and full-length proteins, the progress to date is very promising. The many lessons learned during the experiments described in this Account will help us and others to realize the full catalytic power of DNA.
Figures



References
-
- Flynn-Charlebois A, Wang Y, Prior TK, Rashid I, Hoadley KA, Coppins RL, Wolf AC, Silverman SK. Deoxyribozymes with 2'-5' RNA Ligase Activity. J. Am. Chem. Soc. 2003;125:2444–2454. - PubMed
-
- Flynn-Charlebois A, Prior TK, Hoadley KA, Silverman SK. In Vitro Evolution of an RNA-Cleaving DNA Enzyme into an RNA Ligase Switches the Selectivity from 3'-5' to 2'-5'. J. Am. Chem. Soc. 2003;125:5346–5350. - PubMed
-
- Ricca BL, Wolf AC, Silverman SK. Optimization and Generality of a Small Deoxyribozyme that Ligates RNA. J. Mol. Biol. 2003;330:1015–1025. - PubMed
-
- Semlow DR, Silverman SK. Parallel Selections in Vitro Reveal a Preference for 2'-5' RNA Ligation By Deoxyribozyme-Mediated Opening of a 2',3'-Cyclic Phosphate. J. Mol. Evol. 2005;61:207–215. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

