A non-homogeneous hidden-state model on first order differences for automatic detection of nucleosome positions
- PMID: 19572828
- PMCID: PMC2861327
- DOI: 10.2202/1544-6115.1454
A non-homogeneous hidden-state model on first order differences for automatic detection of nucleosome positions
Abstract
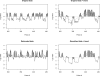
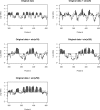
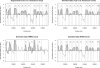
The ability to map individual nucleosomes accurately across genomes enables the study of relationships between dynamic changes in nucleosome positioning/occupancy and gene regulation. However, the highly heterogeneous nature of nucleosome densities across genomes and short linker regions pose challenges in mapping nucleosome positions based on high-throughput microarray data of micrococcal nuclease (MNase) digested DNA. Previous works rely on additional detrending and careful visual examination to detect low-signal nucleosomes, which may exist in a subpopulation of cells. We propose a non-homogeneous hidden-state model based on first order differences of experimental data along genomic coordinates that bypasses the need for local detrending and can automatically detect nucleosome positions of various occupancy levels. Our proposed approach is applicable to both low and high resolution MNase-Chip and MNase-Seq (high throughput sequencing) data, and is able to map nucleosome-linker boundaries accurately. This automated algorithm is also computationally efficient and only requires a simple preprocessing step. We provide several examples illustrating the pitfalls of existing methods, the difficulties of detrending the observed hybridization signals and demonstrate the advantages of utilizing first order differences in detecting nucleosome occupancies via simulations and case studies involving MNase-Chip and MNase-Seq data of nucleosome occupancy in yeast S. cerevisiae.
Figures


















References
-
- Chakravarthy S, Park Y, Chodaparambil J, Edayathumangalam R, Luger K. Structure and dynamic properties of nucleosome core particles, FEBS Letters. 2006;579(4):895–898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
