Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar Typhimurium strains
- PMID: 19573249
- PMCID: PMC2715408
- DOI: 10.1186/1471-2180-9-131
Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar Typhimurium strains
Abstract
Background: Bacterial genomes are mosaic structures composed of genes present in every strain of the same species (core genome), and genes present in some but not all strains of a species (accessory genome). The aim of this study was to compare the genetic diversity of core and accessory genes of a Salmonella enterica subspecies enterica serovar Typhimurium (Typhimurium) population isolated from food-animal and human sources in four regions of Mexico. Multilocus sequence typing (MLST) and macrorestriction fingerprints by pulsed-field gel electrophoresis (PFGE) were used to address the core genetic variation, and genes involved in pathogenesis and antibiotic resistance were selected to evaluate the accessory genome.
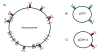
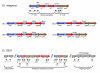
Results: We found a low genetic diversity for both housekeeping and accessory genes. Sequence type 19 (ST19) was supported as the founder genotype of STs 213, 302 and 429. We found a temporal pattern in which the derived ST213 is replacing the founder ST19 in the four geographic regions analyzed and a geographic trend in the number of resistance determinants. The distribution of the accessory genes was not random among chromosomal genotypes. We detected strong associations among the different accessory genes and the multilocus chromosomal genotypes (STs). First, the Salmonella virulence plasmid (pSTV) was found mostly in ST19 isolates. Second, the plasmid-borne betalactamase cmy-2 was found only in ST213 isolates. Third, the most abundant integron, IP-1 (dfrA12, orfF and aadA2), was found only in ST213 isolates. Fourth, the Salmonella genomic island (SGI1) was found mainly in a subgroup of ST19 isolates carrying pSTV. The mapping of accessory genes and multilocus genotypes on the dendrogram derived from macrorestiction fingerprints allowed the establishment of genetic subgroups within the population.
Conclusion: Despite the low levels of genetic diversity of core and accessory genes, the non-random distribution of the accessory genes across chromosomal backgrounds allowed us to discover genetic subgroups within the population. This study provides information about the importance of the accessory genome in generating genetic variability within a bacterial population.
Figures


Similar articles
-
Antimicrobial resistance, plasmid, virulence, multilocus sequence typing and pulsed-field gel electrophoresis profiles of Salmonella enterica serovar Typhimurium clinical and environmental isolates from India.PLoS One. 2018 Dec 12;13(12):e0207954. doi: 10.1371/journal.pone.0207954. eCollection 2018. PLoS One. 2018. PMID: 30540810 Free PMC article.
-
Salmonella enterica serovar Typhimurium virulence-resistance plasmids derived from the pSLT carrying nonconventional class 1 integrons with dfrA12 gene in their variable region and sul3 in the 3' conserved segment.Microb Drug Resist. 2013 Dec;19(6):437-45. doi: 10.1089/mdr.2012.0226. Epub 2013 Jun 29. Microb Drug Resist. 2013. PMID: 23808958
-
Detection of multidrug-resistant Salmonella enterica serovar typhimurium phage types DT102, DT104, and U302 by multiplex PCR.J Clin Microbiol. 2006 Jul;44(7):2354-8. doi: 10.1128/JCM.00171-06. J Clin Microbiol. 2006. PMID: 16825349 Free PMC article.
-
Presence of integrons and their correlation with multidrug resistance in Salmonella enterica serovar Typhimurium: Exploratory systematic review.Biomedica. 2024 May 30;44(2):258-276. doi: 10.7705/biomedica.6816. Biomedica. 2024. PMID: 39088536 Free PMC article. English, Spanish.
-
The Salmonella enterica pan-genome.Microb Ecol. 2011 Oct;62(3):487-504. doi: 10.1007/s00248-011-9880-1. Epub 2011 Jun 4. Microb Ecol. 2011. PMID: 21643699 Free PMC article. Review.
Cited by
-
Antimicrobial resistance, plasmid, virulence, multilocus sequence typing and pulsed-field gel electrophoresis profiles of Salmonella enterica serovar Typhimurium clinical and environmental isolates from India.PLoS One. 2018 Dec 12;13(12):e0207954. doi: 10.1371/journal.pone.0207954. eCollection 2018. PLoS One. 2018. PMID: 30540810 Free PMC article.
-
Multilocus sequence typing as a replacement for serotyping in Salmonella enterica.PLoS Pathog. 2012;8(6):e1002776. doi: 10.1371/journal.ppat.1002776. Epub 2012 Jun 21. PLoS Pathog. 2012. PMID: 22737074 Free PMC article.
-
Characterization of Salmonella enterica isolates causing bacteremia in Lima, Peru, using multiple typing methods.PLoS One. 2017 Dec 21;12(12):e0189946. doi: 10.1371/journal.pone.0189946. eCollection 2017. PLoS One. 2017. PMID: 29267322 Free PMC article.
-
Complete Genome Sequencing of a Multidrug-Resistant and Human-Invasive Salmonella enterica Serovar Typhimurium Strain of the Emerging Sequence Type 213 Genotype.Genome Announc. 2015 Jun 18;3(3):e00663-15. doi: 10.1128/genomeA.00663-15. Genome Announc. 2015. PMID: 26089426 Free PMC article.
-
Association of Neisseria gonorrhoeae Plasmids With Distinct Lineages and The Economic Status of Their Country of Origin.J Infect Dis. 2020 Nov 9;222(11):1826-1836. doi: 10.1093/infdis/jiaa003. J Infect Dis. 2020. PMID: 32163577 Free PMC article.
References
-
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae : implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

