Discovering rules for protein-ligand specificity using support vector inductive logic programming
- PMID: 19574295
- PMCID: PMC3913550
- DOI: 10.1093/protein/gzp035
Discovering rules for protein-ligand specificity using support vector inductive logic programming
Abstract
Structural genomics initiatives are rapidly generating vast numbers of protein structures. Comparative modelling is also capable of producing accurate structural models for many protein sequences. However, for many of the known structures, functions are not yet determined, and in many modelling tasks, an accurate structural model does not necessarily tell us about function. Thus, there is a pressing need for high-throughput methods for determining function from structure. The spatial arrangement of key amino acids in a folded protein, on the surface or buried in clefts, is often the determinants of its biological function. A central aim of molecular biology is to understand the relationship between such substructures or surfaces and biological function, leading both to function prediction and to function design. We present a new general method for discovering the features of binding pockets that confer specificity for particular ligands. Using a recently developed machine-learning technique which couples the rule-discovery approach of inductive logic programming with the statistical learning power of support vector machines, we are able to discriminate, with high precision (90%) and recall (86%) between pockets that bind FAD and those that bind NAD on a large benchmark set given only the geometry and composition of the backbone of the binding pocket without the use of docking. In addition, we learn rules governing this specificity which can feed into protein functional design protocols. An analysis of the rules found suggests that key features of the binding pocket may be tied to conformational freedom in the ligand. The representation is sufficiently general to be applicable to any discriminatory binding problem. All programs and data sets are freely available to non-commercial users at http://www.sbg.bio.ic.ac.uk/svilp_ligand/.
Figures
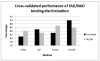
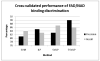
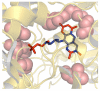
Similar articles
-
A general approach for developing system-specific functions to score protein-ligand docked complexes using support vector inductive logic programming.Proteins. 2007 Dec 1;69(4):823-31. doi: 10.1002/prot.21782. Proteins. 2007. PMID: 17910057
-
Conformational diversity of ligands bound to proteins.J Mol Biol. 2006 Mar 3;356(4):928-44. doi: 10.1016/j.jmb.2005.12.012. Epub 2005 Dec 20. J Mol Biol. 2006. PMID: 16405908
-
Adenine recognition: a motif present in ATP-, CoA-, NAD-, NADP-, and FAD-dependent proteins.Proteins. 2001 Aug 15;44(3):282-91. doi: 10.1002/prot.1093. Proteins. 2001. PMID: 11455601
-
Sequence-structure analysis of FAD-containing proteins.Protein Sci. 2001 Sep;10(9):1712-28. doi: 10.1110/ps.12801. Protein Sci. 2001. PMID: 11514662 Free PMC article. Review.
-
Importance of molecular computer modeling in anticancer drug development.J BUON. 2007 Sep;12 Suppl 1:S101-18. J BUON. 2007. PMID: 17935268 Review.
Cited by
-
Sunsetting Binding MOAD with its last data update and the addition of 3D-ligand polypharmacology tools.Sci Rep. 2023 Feb 21;13(1):3008. doi: 10.1038/s41598-023-29996-w. Sci Rep. 2023. PMID: 36810894 Free PMC article.
-
LIMLE, a new molecule over-expressed following activation, is involved in the stimulatory properties of dendritic cells.PLoS One. 2014 Apr 4;9(4):e93894. doi: 10.1371/journal.pone.0093894. eCollection 2014. PLoS One. 2014. PMID: 24705920 Free PMC article.
-
Knowledge discovery in variant databases using inductive logic programming.Bioinform Biol Insights. 2013 Mar 18;7:119-31. doi: 10.4137/BBI.S11184. Print 2013. Bioinform Biol Insights. 2013. PMID: 23589683 Free PMC article.
-
Homology modeling and structural comparison of leucine rich repeats of Toll like receptors 1-10 of ruminants.J Mol Model. 2013 Sep;19(9):3863-74. doi: 10.1007/s00894-013-1871-3. Epub 2013 Jun 28. J Mol Model. 2013. PMID: 23812948
References
-
- Amini A, Lodhi H, Muggleton SH, Sternberg MJE. J. Chem. Inf. Model. 2007;47(3):998–1006. - PubMed
-
- Amini A, Shrimpton PJ, Muggleton SH, Sternberg MJE. Proteins: Struct., Funct., Bioinf. 2007;69(4):823–831. - PubMed
-
- Brenner SE. Nat Rev Genet. 2001;2(10):801–809. - PubMed
-
- Burley SK, Almo SC, Bonanno JB, Capel M, Chance MR, Gaasterland T, Lin D, Sali A, Studier FW, Swaminathan S. Nat. Genet. 1999;23(2):151–157. - PubMed
-
- Baker D, Sali A. Science. 2001;294(5540):93–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

