The modular architecture of meningococcal factor H-binding protein
- PMID: 19574307
- PMCID: PMC2859308
- DOI: 10.1099/mic.0.029876-0
The modular architecture of meningococcal factor H-binding protein
Abstract
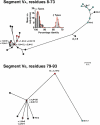
Meningococcal factor H binding protein (fHbp) is a promising vaccine antigen that binds the human complement downregulatory molecule factor H (fH), and this binding enhances the survival of the organism in serum. Based on sequence variability of the entire protein, fHbp has been divided into three variant groups or two subfamilies. Here, we present evidence based on phylogenetic analysis of 70 unique fHbp amino acid sequences that the molecular architecture is modular. From sequences of natural chimeras we identified blocks of two to five invariant residues that flanked five modular variable segments. Although overall, 46 % of the fHbp amino acids were invariant, based on a crystal structure, the invariant blocks that flanked the modular variable segments clustered on the membrane surface containing the amino-terminal lipid anchor, while the remaining invariant residues were located throughout the protein. Each of the five modular variable segments could be classified into one of two types, designated alpha or beta, based on homology with segments encoded by variant 1 or 3 fHbp genes, respectively. Forty of the fHbps (57 %) comprised only alpha (n=33) or beta (n=7) type segments. The remaining 30 proteins (43 %) were chimeras and could be classified into one of four modular groups. These included all 15 proteins assigned to the previously described variant 2 in subfamily A. The modular segments of one chimeric modular group had 96 % amino acid identity with those of fHbp orthologs in Neisseria gonorrhoeae. Collectively, the data suggest that recombination between Neisseria meningitidis and N. gonorrhoeae progenitors generated a family of modular, antigenically diverse meningococcal fHbps.
Figures



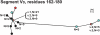

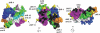
References
-
- Arreaza, L., Alcala, B., Salcedo, C., de la Fuente, L. & Vazquez, J. A. (2003). Dynamics of the penA gene in serogroup C meningococcal strains. J Infect Dis 187, 1010–1014. - PubMed
-
- Bambini, S., Muzzi, A., Olcen, P., Rappuoli, R., Pizza, M. & Comanducci, M. (2009). Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 27, 2794–2803. - PubMed
-
- Beernink, P. T., Welsch, J. A., Harrison, L. H., Leipus, A., Kaplan, S. L. & Granoff, D. M. (2007). Prevalence of factor H-binding protein variants and NadA among meningococcal group B isolates from the United States: implications for the development of a multicomponent group B vaccine. J Infect Dis 195, 1472–1479. - PMC - PubMed
-
- Beernink, P. T., Welsch, J. A., Bar-Lev, M., Koeberling, O., Comanducci, M. & Granoff, D. M. (2008). Fine antigenic specificity and cooperative bactericidal activity of monoclonal antibodies directed at the meningococcal vaccine candidate, factor H-binding protein. Infect Immun 76, 4232–4240. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

