Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
- PMID: 19574954
- PMCID: PMC2735180
- DOI: 10.1038/emboj.2009.184
Insights into complement convertase formation based on the structure of the factor B-cobra venom factor complex
Abstract
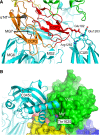
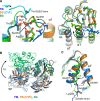
Immune protection by the complement system critically depends on assembly of C3 convertases on the surface of pathogens and altered host cells. These short-lived protease complexes are formed through pro-convertases, which for the alternative pathway consist of the complement component C3b and the pro-enzyme factor B (FB). Here, we present the crystal structure at 2.2-A resolution, small-angle X-ray scattering and electron microscopy (EM) data of the pro-convertase formed by human FB and cobra venom factor (CVF), a potent homologue of C3b that generates more stable convertases. FB is loaded onto CVF through its pro-peptide Ba segment by specific contacts, which explain the specificity for the homologous C3b over the native C3 and inactive products iC3b and C3c. The protease segment Bb binds the carboxy terminus of CVF through the metal-ion dependent adhesion site of the Von Willebrand factor A-type domain. A possible dynamic equilibrium between a 'loading' and 'activation' state of the pro-convertase may explain the observed difference between the crystal structure of CVFB and the EM structure of C3bB. These insights into formation of convertases provide a basis for further development of complement therapeutics.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



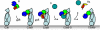
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 - PubMed
-
- Bhattacharya AA, Lupher ML Jr, Staunton DE, Liddington RC (2004) Crystal structure of the A domain from complement factor B reveals an integrin-like open conformation. Structure (Camb) 12: 371–378 - PubMed
-
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 - PubMed
-
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

