The biogenesis and function of PIWI proteins and piRNAs: progress and prospect
- PMID: 19575643
- PMCID: PMC2780330
- DOI: 10.1146/annurev.cellbio.24.110707.175327
The biogenesis and function of PIWI proteins and piRNAs: progress and prospect
Abstract
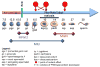
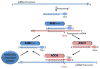
The evolutionarily conserved Argonaute/PIWI (AGO/PIWI, also known as PAZ-PIWI domain or PPD) family of proteins is crucial for the biogenesis and function of small noncoding RNAs (ncRNAs). This family can be divided into AGO and PIWI subfamilies. The AGO proteins are ubiquitously present in diverse tissues. They bind to small interfering RNAs (siRNAs) and microRNAs (miRNAs). In contrast, the PIWI proteins are predominantly present in the germline and associate with a novel class of small RNAs known as PIWI-interacting RNAs (piRNAs). Tens of thousands of piRNA species, typically 24-32 nucleotide (nt) long, have been found in mammals, zebrafish, and Drosophila. Most piRNAs appear to be generated from a small number of long single-stranded RNA precursors that are often encoded by repetitive intergenic sequences in the genome. PIWI proteins play crucial roles during germline development and gametogenesis of many metazoan species, from germline determination and germline stem cell (GSC) maintenance to meiosis, spermiogenesis, and transposon silencing. These diverse functions may involve piRNAs and may be achieved via novel mechanisms of epigenetic and posttranscriptional regulation.
Figures


References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442(7099):203–207. - PubMed
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5(2):337–350. - PubMed
-
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11(13):1017–1027. - PubMed
-
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316(5825):744–747. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

