Small RNAs and their roles in plant development
- PMID: 19575669
- PMCID: PMC5135726
- DOI: 10.1146/annurev.cellbio.042308.113417
Small RNAs and their roles in plant development
Abstract
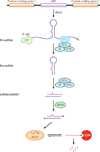
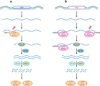
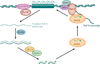
Small RNAs of 20-30 nucleotides guide regulatory processes at the DNA or RNA level in a wide range of eukaryotic organisms. Many, although not all, small RNAs are processed from double-stranded RNAs or single-stranded RNAs with local hairpin structures by RNase III enzymes and are loaded into argonaute-protein-containing effector complexes. Many eukaryotic organisms have evolved multiple members of RNase III and the argonaute family of proteins to accommodate different classes of small RNAs with specialized molecular functions. Some small RNAs cause transcriptional gene silencing by guiding heterochromatin formation at homologous loci, whereas others lead to posttranscriptional gene silencing through mRNA degradation or translational inhibition. Small RNAs are not only made from and target foreign nucleic acids such as viruses and transgenes, but are also derived from endogenous loci and regulate a multitude of developmental and physiological processes. Here I review the biogenesis and function of three major classes of endogenous small RNAs in plants: microRNAs, trans-acting siRNAs, and heterochromatic siRNAs, with an emphasis on the roles of these small RNAs in developmental regulation.
Figures

References
-
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. - PubMed
-
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7 . Curr. Biol. 2006;16:927–932. - PubMed
-
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. - PubMed
-
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. - PubMed
-
- Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004;36:1282–1290. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

