Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases
- PMID: 19575810
- PMCID: PMC2721837
- DOI: 10.1186/1472-6807-9-42
Conservation of structure and activity in Plasmodium purine nucleoside phosphorylases
Abstract
Background: Purine nucleoside phosphorylase (PNP) is central to purine salvage mechanisms in Plasmodium parasites, the causative agents of malaria. Most human malaria results from infection either by Plasmodium falciparum (Pf), the deadliest form of the parasite, or by the widespread Plasmodium vivax (Pv). Whereas the PNP enzyme from Pf has previously been studied in detail, despite the prevalence of Pv little is known about many of the key metabolic enzymes from this parasite, including PvPNP.
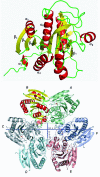
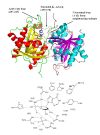
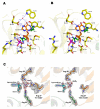
Results: The crystal structure of PvPNP is described and is seen to have many features in common with the previously reported structure of PfPNP. In particular, the composition and conformations of the active site regions are virtually identical. The crystal structure of a complex of PfPNP co-crystallised with inosine and arsenate is also described, and is found to contain a mixture of products and reactants - hypoxanthine, ribose and arsenate. The ribose C1' in this hybrid complex lies close to the expected point of symmetry along the PNP reaction coordinate, consistent with a conformation between the transition and product states. These two Plasmodium PNP structures confirm the similarity of structure and mechanism of these enzymes, which are also confirmed in enzyme kinetic assays using an array of substrates. These reveal an unusual form of substrate activation by 2'-deoxyinosine of PvPNP, but not PfPNP.
Conclusion: The close similarity of the Pf and Pv PNP structures allows characteristic features to be identified that differentiate the Apicomplexa PNPs from the human host enzyme. This similarity also suggests there should be a high level of cross-reactivity for compounds designed to inhibit either of these molecular targets. However, despite these similarities, there are also small differences in the activities of the two Plasmodium enzymes.
Figures
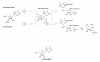


Similar articles
-
Energetic mapping of transition state analogue interactions with human and Plasmodium falciparum purine nucleoside phosphorylases.J Biol Chem. 2005 Aug 26;280(34):30320-8. doi: 10.1074/jbc.M505033200. Epub 2005 Jun 16. J Biol Chem. 2005. PMID: 15961383
-
Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases.Biochemistry. 2004 Feb 17;43(6):1458-68. doi: 10.1021/bi0359123. Biochemistry. 2004. PMID: 14769022
-
The purine nucleoside phosphorylase from Trichomonas vaginalis is a homologue of the bacterial enzyme.Biochemistry. 2002 Aug 20;41(33):10382-9. doi: 10.1021/bi026025n. Biochemistry. 2002. PMID: 12173924
-
Targeting Plasmodium falciparum purine salvage enzymes: a look at structure-based drug development.Infect Disord Drug Targets. 2010 Jun;10(3):191-9. doi: 10.2174/187152610791163408. Infect Disord Drug Targets. 2010. PMID: 20480551 Review.
-
Transition states and inhibitors of the purine nucleoside phosphorylase family.Curr Top Med Chem. 2005;5(13):1237-58. doi: 10.2174/156802605774463088. Curr Top Med Chem. 2005. PMID: 16305529 Review.
Cited by
-
Metabolomics Analysis of Hippocampus and Cortex in a Rat Model of Traumatic Brain Injury in the Subacute Phase.Front Neurosci. 2020 Sep 4;14:876. doi: 10.3389/fnins.2020.00876. eCollection 2020. Front Neurosci. 2020. PMID: 33013291 Free PMC article.
-
Lys48 ubiquitination during the intraerythrocytic cycle of the rodent malaria parasite, Plasmodium chabaudi.PLoS One. 2017 Jun 12;12(6):e0176533. doi: 10.1371/journal.pone.0176533. eCollection 2017. PLoS One. 2017. PMID: 28604779 Free PMC article.
-
The Potential of Secondary Metabolites from Plants as Drugs or Leads against Protozoan Neglected Diseases-Part III: In-Silico Molecular Docking Investigations.Molecules. 2016 Oct 19;21(10):1389. doi: 10.3390/molecules21101389. Molecules. 2016. PMID: 27775577 Free PMC article. Review.
-
Arsenate replacing phosphate: alternative life chemistries and ion promiscuity.Biochemistry. 2011 Feb 22;50(7):1128-34. doi: 10.1021/bi200002a. Epub 2011 Jan 31. Biochemistry. 2011. PMID: 21214261 Free PMC article. Review.
-
Identification and structural validation of purine nucleoside phosphorylase from Plasmodium falciparum as a target of MMV000848.J Biol Chem. 2024 Jan;300(1):105586. doi: 10.1016/j.jbc.2023.105586. Epub 2023 Dec 21. J Biol Chem. 2024. PMID: 38141766 Free PMC article.
References
-
- Ting LM, Shi W, Lewandowicz A, Singh V, Mwakingwe A, Birck MR, Ringia EA, Bench G, Madrid DC, Tyler PC, et al. Targeting a novel Plasmodium falciparum purine recycling pathway with specific immucillins. J Biol Chem. 2005;280:9547–9554. - PubMed
-
- Lewandowicz A, Schramm VL. Transition state analysis for human and Plasmodium falciparum purine nucleoside phosphorylases. Biochemistry. 2004;43:1458–1468. - PubMed
-
- Kline PC, Schramm VL. Pre-Steady-State Transition-State Analysis of the Hydrolytic Reaction Catalyzed by Purine Nucleoside Phosphorylase. Biochemistry. 1995;34:1153–1162. - PubMed
-
- Fedorov A, Shi W, Kicska G, Fedorov E, Tyler PC, Furneaux RH, Hanson JC, Gainsford GJ, Larese JZ, Schramm VL, et al. Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis. Biochemistry. 2001;40:853–860. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

