Gene Ontology and the annotation of pathogen genomes: the case of Candida albicans
- PMID: 19577928
- PMCID: PMC3907193
- DOI: 10.1016/j.tim.2009.04.007
Gene Ontology and the annotation of pathogen genomes: the case of Candida albicans
Abstract
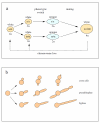
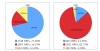
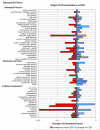
The Gene Ontology (GO) is a structured controlled vocabulary developed to describe the roles and locations of gene products in a consistent manner and in a way that can be shared across organisms. The unicellular fungus Candida albicans is similar in many ways to the model organism Saccharomyces cerevisiae but, as both a commensal and a pathogen of humans, differs greatly in its lifestyle. With an expanding at-risk population of immunosuppressed patients, increased use of invasive medical procedures, the increasing prevalence of drug resistance and the emergence of additional Candida species as serious pathogens, it has never been more crucial to improve our understanding of Candida biology to guide the development of better treatments. In this brief review, we examine the importance of GO in the annotation of C. albicans gene products, with a focus on those involved in pathogenesis. We also discuss how sequence information combined with GO facilitates the transfer of knowledge across related species and the challenges and opportunities that such an approach presents.
Figures

References
-
- Schelenz S. Management of candidiasis in the intensive care unit. The Journal of antimicrobial chemotherapy. 2008;61(Suppl 1):i31–34. - PubMed
-
- d'Enfert C. Biofilms and their role in the resistance of pathogenic Candida to antifungal agents. Current drug targets. 2006;7:465–470. - PubMed
-
- Lewis K. Multidrug tolerance of biofilms and persister cells. Current topics in microbiology and immunology. 2008;322:107–131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

