Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii
- PMID: 19578403
- PMCID: PMC2697382
- DOI: 10.1371/journal.pgen.1000546
Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii
Abstract
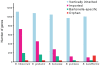
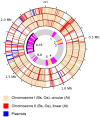
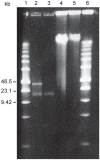
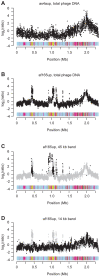
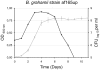
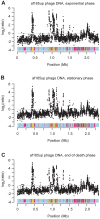
The genus Bartonella comprises facultative intracellular bacteria adapted to mammals, including previously recognized and emerging human pathogens. We report the 2,341,328 bp genome sequence of Bartonella grahamii, one of the most prevalent Bartonella species in wild rodents. Comparative genomics revealed that rodent-associated Bartonella species have higher copy numbers of genes for putative host-adaptability factors than the related human-specific pathogens. Many of these gene clusters are located in a highly dynamic region of 461 kb. Using hybridization to a microarray designed for the B. grahamii genome, we observed a massive, putatively phage-derived run-off replication of this region. We also identified a novel gene transfer agent, which packages the bacterial genome, with an over-representation of the amplified DNA, in 14 kb pieces. This is the first observation associating the products of run-off replication with a gene transfer agent. Because of the high concentration of gene clusters for host-adaptation proteins in the amplified region, and since the genes encoding the gene transfer agent and the phage origin are well conserved in Bartonella, we hypothesize that these systems are driven by selection. We propose that the coupling of run-off replication with gene transfer agents promotes diversification and rapid spread of host-adaptability factors, facilitating host shifts in Bartonella.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Dehio C. Bartonella interactions with endothelial cells and erythrocytes. Trends Microbiol. 2001;9:279–85. - PubMed
-
- Saenz HL, Engel P, Stoeckli MC, Lanz C, Raddatz G, et al. Genomic analysis of Bartonella identifies type IV secretion systems as host adaptability factors. Nat Genet. 2007;39:1469–76. - PubMed
-
- Schulein R, Dehio C. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol Microbiol. 2002;46:1053–67. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

