The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species
- PMID: 19578431
- PMCID: PMC2699109
- DOI: 10.1371/journal.pcbi.1000431
The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species
Abstract
The Gene Ontology (GO) is a collaborative effort that provides structured vocabularies for annotating the molecular function, biological role, and cellular location of gene products in a highly systematic way and in a species-neutral manner with the aim of unifying the representation of gene function across different organisms. Each contributing member of the GO Consortium independently associates GO terms to gene products from the organism(s) they are annotating. Here we introduce the Reference Genome project, which brings together those independent efforts into a unified framework based on the evolutionary relationships between genes in these different organisms. The Reference Genome project has two primary goals: to increase the depth and breadth of annotations for genes in each of the organisms in the project, and to create data sets and tools that enable other genome annotation efforts to infer GO annotations for homologous genes in their organisms. In addition, the project has several important incidental benefits, such as increasing annotation consistency across genome databases, and providing important improvements to the GO's logical structure and biological content.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
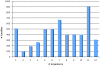

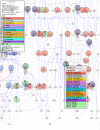
References
-
- Rhee SY, Wood V, Dolinski K, Draghici S. Use and misuse of the gene ontology annotations. Nat Rev Genet. 2008;9:509–515. - PubMed
Publication types
MeSH terms
Grants and funding
- P41 HG002659/HG/NHGRI NIH HHS/United States
- U41 HG002273/HG/NHGRI NIH HHS/United States
- HL64541/HL/NHLBI NIH HHS/United States
- P41 HG000330/HG/NHGRI NIH HHS/United States
- U41 HG002223/HG/NHGRI NIH HHS/United States
- P41 HG002659-06/HG/NHGRI NIH HHS/United States
- P41 HG02223/HG/NHGRI NIH HHS/United States
- U24 HG001315/HG/NHGRI NIH HHS/United States
- R01 GM064426/GM/NIGMS NIH HHS/United States
- HD033745/HD/NICHD NIH HHS/United States
- P41 HG002273/HG/NHGRI NIH HHS/United States
- U24 HG002659/HG/NHGRI NIH HHS/United States
- U41 HG001315/HG/NHGRI NIH HHS/United States
- U24 HG002223/HG/NHGRI NIH HHS/United States
- R01 HL064541/HL/NHLBI NIH HHS/United States
- SP/07/007/23671/BHF_/British Heart Foundation/United Kingdom
- P41 HG001315/HG/NHGRI NIH HHS/United States
- G0500293/MRC_/Medical Research Council/United Kingdom
- P41 HG002223/HG/NHGRI NIH HHS/United States
- R01 HD033745/HD/NICHD NIH HHS/United States
- U41 HG002659/HG/NHGRI NIH HHS/United States
- U24 GM07790/GM/NIGMS NIH HHS/United States
- R01 HG002273/HG/NHGRI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- T32 GM007790/GM/NIGMS NIH HHS/United States
- HG00022/HG/NHGRI NIH HHS/United States
- GM64426/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

