The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output
- PMID: 19578433
- PMCID: PMC2698984
- DOI: 10.1371/journal.ppat.1000496
The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output
Abstract
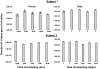
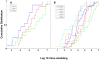
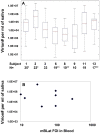
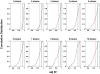
To develop more detailed models of EBV persistence we have studied the dynamics of virus shedding in healthy carriers. We demonstrate that EBV shedding into saliva is continuous and rapid such that the virus level is replaced in < or =2 minutes, the average time that a normal individual swallows. Thus, the mouth is not a reservoir of virus but a conduit through which a continuous flow stream of virus passes in saliva. Consequently, virus is being shed at a much higher rate than previously thought, a level too high to be accounted for by replication in B cells in Waldeyer's ring alone. Virus shedding is relatively stable over short periods (hours-days) but varies through 3.5 to 5.5 logs over longer periods, a degree of variation that also cannot be accounted for solely by replication in B cells. This variation means, contrary to what is generally believed, that the definition of high and low shedder is not so much a function of variation between individuals but within individuals over time. The dynamics of shedding describe a process governing virus production that is occurring independently < or =3 times at any moment. This process grows exponentially and is then randomly terminated. We propose that these dynamics are best explained by a model where single B cells sporadically release virus that infects anywhere from 1 to 5 epithelial cells. This infection spreads at a constant exponential rate and is terminated randomly, resulting in infected plaques of epithelial cells ranging in size from 1 to 10(5) cells. At any one time there are a very small number (< or =3) of plaques. We suggest that the final size of these plaques is a function of the rate of infectious spread within the lymphoepithelium which may be governed by the structural complexity of the tissue but is ultimately limited by the immune response.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Kieff E, Rickinson AB. Epstein-Barr Virus and Its Replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2654.
-
- Thorley-Lawson D. EBV persistence and latent infection in vivo. In: ES R, editor. Epstein-Barr Virus. 1st ed. Norfolk, England: Caister Academic Press; 2005. pp. 349.
-
- Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. - PubMed
-
- Rickinson AB, Kieff E. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2700.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

